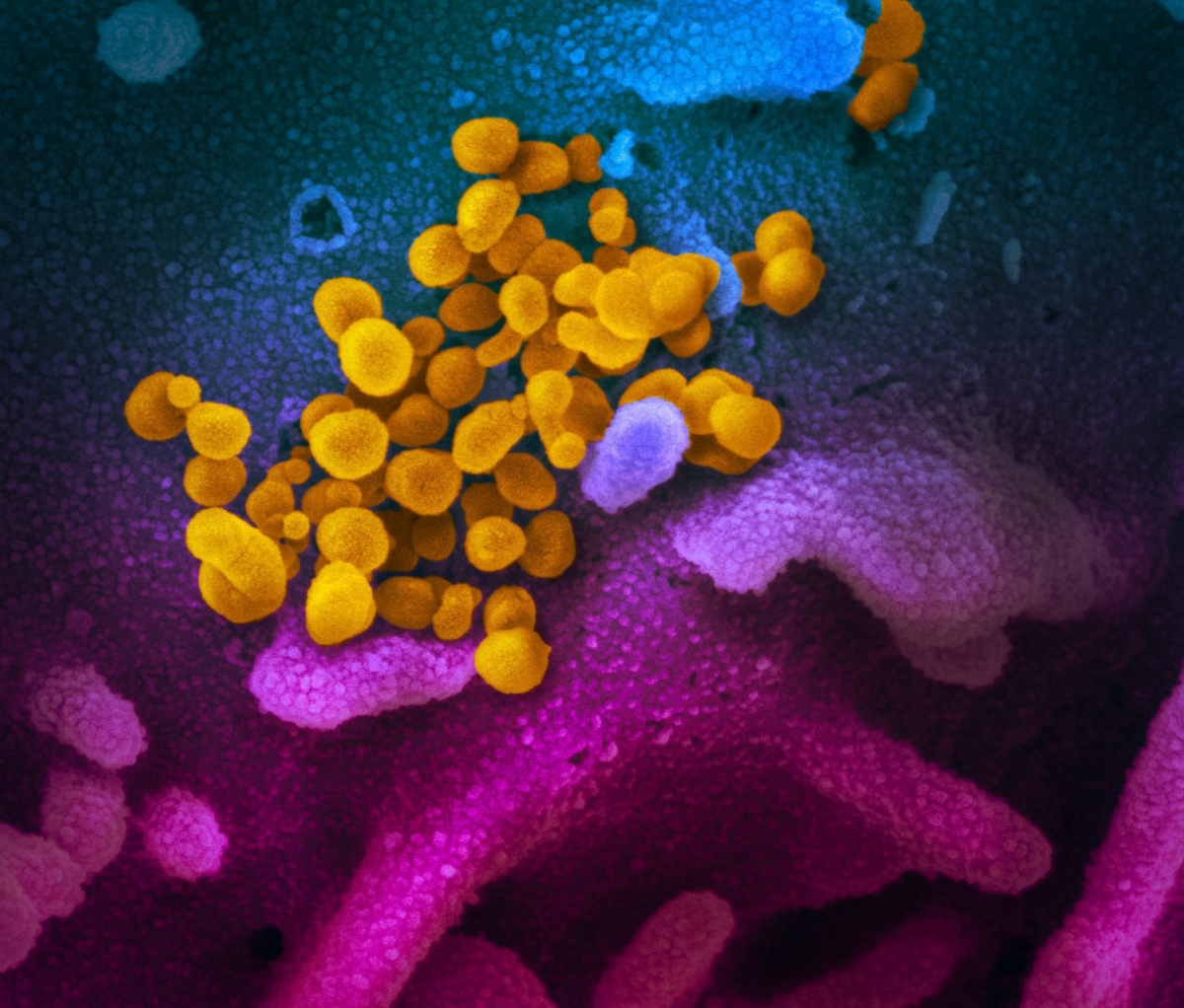
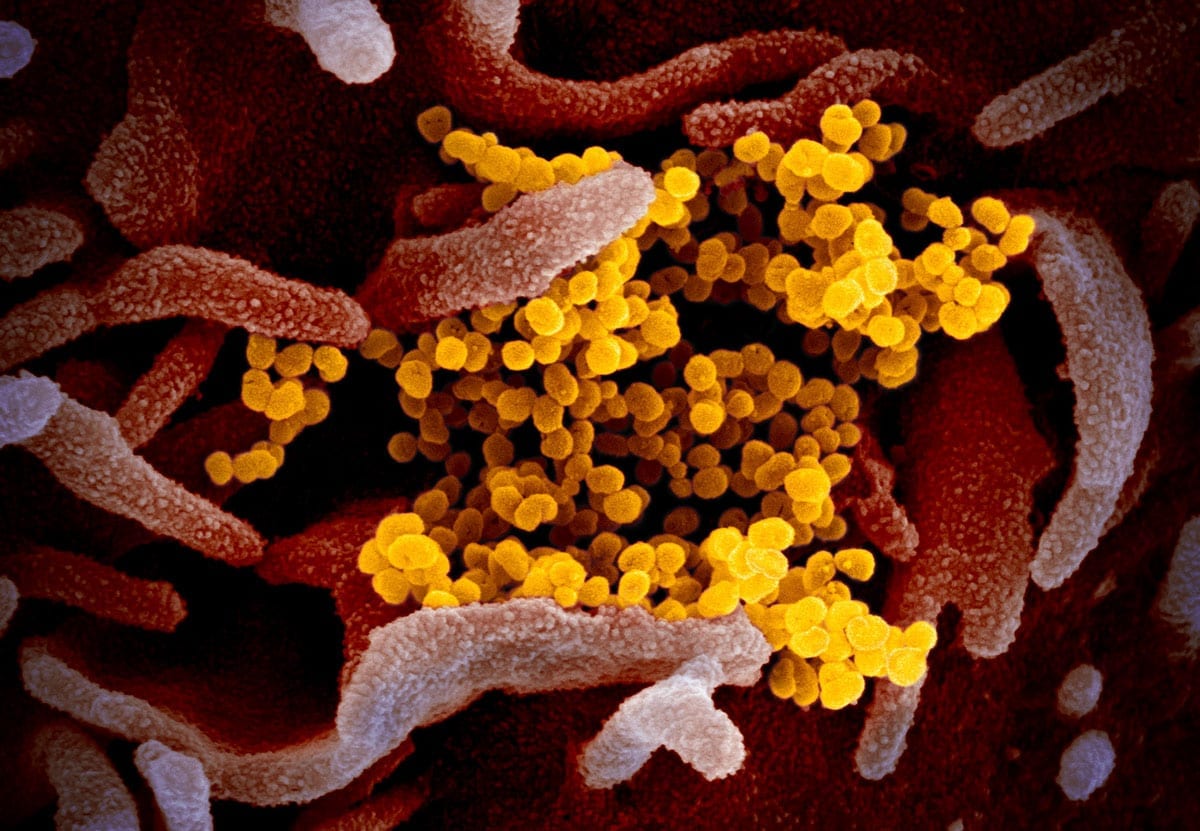
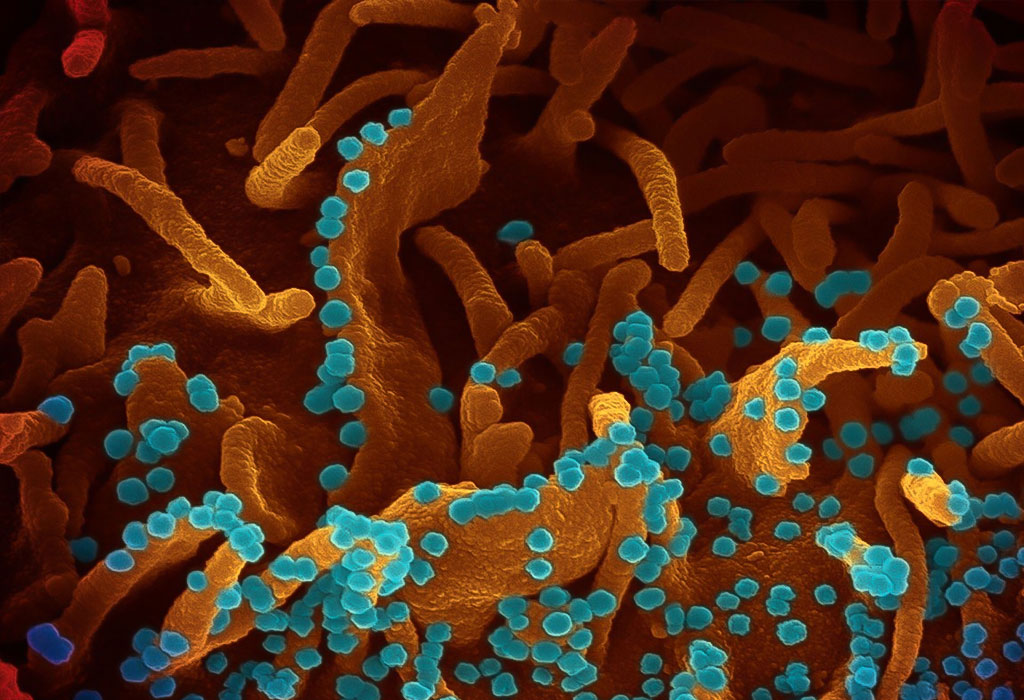
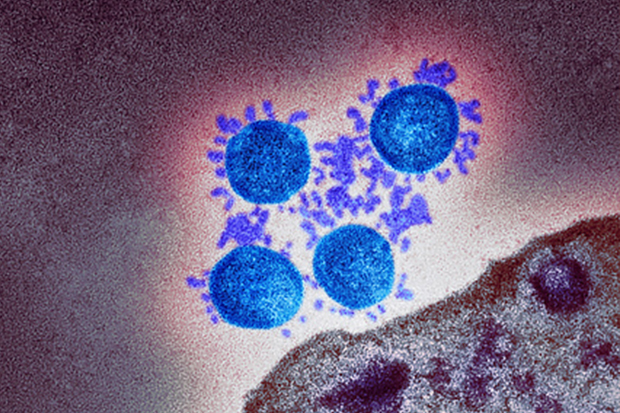



Coloured transmission electron micrograph (TEM) of a SARS-CoV-2 coronavirus particle isolated from a UK case of the disease Covid-19.
Vir: https://publichealthmatters.blog.gov.uk/2021/03/11/the-siren-study-answering-the-big-questions/
*
Vlada RS nov 2021: Virus SARS-CoV-2 so velikokrat neodvisno izolirali (tudi v Sloveniji), natančno določili zaporedje njegovega genoma, virus so velikokrat analizirali z elektronskim mikroskopom (tudi v laboratorijih Medicinske fakultete v Ljubljani), dokazali so, da se virus lahko razmnožuje v človeških celicah, ter z izoliranim virusom okužili eksperimentalne živali in prostovoljce
https://www.gov.si/assets/ministrstva/MZ/DOKUMENTI/Koronavirus/FAQ_Kaj_vemo_o_covid19_v1-v2.pdf
*
IZOLACIJA VIRUSA SARS CoV-2 V LJUBLJANI-
do oktobra 2021 je bilo v Sloveniji 60 izolacij virusa SARS CoV-2, do januarja 2022 pa več kot 70. Objavljamo odgovor vodilne slovenske virologinje na vprašanje zainteresirane javnosti o izolaciji virusa z dne 11.1. 2022 ter odlomek članka z dne 31. 10. 2021
ZEFO, Zavezništvo Empowerment41 Alliance - inštitut za človeka in okolje je januarja 2022 pisal več slovenskim strokovnjakom in jih prosil za dokazila o obstoju virusa SARS CoV-2. Od Akad. prof. dr. Tatjana Avšič-Županc, univ. dipl. biol., Specialistke medicinske mikrobiologije so prejeli naslednji odgovor:
"Kot virolog vama bom odgovorila le na 1. vprašanje o vajinem dvomu, da virusa do zdaj še niso izolirali?
Virus SARS-CoV-2 so zdaj izolirali v številnih laboratorijih po svetu, vključno z našim laboratorijem na IMI-MF. Namreč v sklopu našega inštituta vodim laboratorij 3. stopnje biološke varnosti, kjer lahko pod varnimi pogoji za delavce in okolje delamo z mikroorganizmi 3. stopnje biološke nevarnosti, kamor sodi tudi virus SARS-CoV-2. Do zdaj smo v našem laboratoriju uspešno izolirali virus SARS-CoV-2 več kot 70 X. Viruse izoliramo zaradi različnih potreb klinikov in predvsem z namenom raziskovanja bioloških in antigenskih lastnosti virusa, ki jih lahko proučujemo le v z živim virusom. Tako imamo izolirane viruse iz nosno žrelnih brisov bolnikov, iz aspiratov traheje bolnikov, ki so na respiratorjih, iz pljučnega tkiva in tkiva srca bolnikov, ki so umrli zaradi covida, iz posteljice porodnice s covidom. Izolirane imamo tudi vse zaskrbljujoče koronavirusne genetske različice (Alfa, Beta, Gama, Delta, Omikron in druge..)."
Akad. prof. dr. Tatjana Avšič-Županc, univ. dipl. biol.,
Specialist medicinske mikrobiologije
Prof. dr. Tatjana Avšič Županc je največja slovenska strokovnjakinja za viruse. Je vodja laboratorija za diagnostiko zoonoz (bolezni, ki se prenašajo z živali na človeka) na Inštitutu za mikrobiologijo in imunologijo in profesorica na ljubljanski Medicinski fakulteti. Je članica SAZU in edina Slovenka, ki je dobila priložnost poimenovati nov virus, ki ga je odkrila.
Med ljudmi se še vedno pojavljajo dvomi o obstoju virusa. Vi ste verjetno najbolj primerna oseba, ki jim lahko ponudi dokaze.
"Virus vsekakor obstaja. Trenutno imamo na inštitutu 60 izolatov virusa SARS-CoV-2, ki nam jih je uspelo osamiti iz različnih kliničnih vzorcev bolnikov ali umrlih zaradi covida-19 v Sloveniji. Izolirali smo tudi vse pomembne genetske različice virusa. S pomočjo elektronskega mikroskopa lahko virus SARS-CoV-2 vidimo tudi neposredno, denimo v pljučnem tkivu umrlih bolnikov. Tam ga je res veliko."
Vir: https://n1info.si/pogl.../intervju-virologinja-avsic-zupanc/
P.s: Medicinska fakulteta tudi prodaja virus SARS CoV-2: 1ml= 2000 EUR: https://www.european-virus-archive.com/.../sars-cov-2...
Demant trditve dr. Kaufmanna, da koronavirus SARS CoV-2 ni bil izoliran in da to ni virus, ampak toksin-eksosom. Če bi to bili eksosomi, se ne bi v celicah "in vitro" razmnoževali, tako kot koronavirusi SARS CoV-2, ampak bi se z večanjem celične kulture razredčili, saj se eksosomi ne razmnožujejo:
*
Več o izolaciji SARS CoV-2 in demant trditev dr. Kaufmanna in dr. Cowana, da virus ni bil izoliran:
https://www.youtube.com/watch?v=2tU5OnaTHcE
Ta slika optičnega elektronskega mikroskopa prikazuje SARS-CoV-2 (rumena) - znan tudi kot 2019-nCoV, virus, ki povzroča COVID-19 - izoliran od pacienta v ZDA, ki izhaja iz površine celic (roza), gojenih v laboratoriju. Zasluga: NIAID-RML
Raziskovalka RML dr. Emmie de Wit je v okviru svojih študij priskrbela vzorce virusa. Mikroskopinja Elizabeth Fischer je izdelala slike, urad za vizualno medicino RML pa jih je digitalno obarval.
The images are available free to the public on the NIAID Flickr page. NIAID asks all who use the images to please credit NIAID-RML.
Ta slika optičnega elektronskega mikroskopa prikazuje SARS-CoV-2 (rumena) - znan tudi kot 2019-nCoV, virus, ki povzroča COVID-19 - izoliran od pacienta v ZDA, ki izhaja iz površine gojenih celic (modra / roza) v laboratoriju. Zasluga: NIAID-RML
Nov koronavirus SARS-CoV-2 Ta posnetek elektronskega mikroskopa prikazuje SARS-CoV-2 - znan tudi kot 2019-nCoV, virus, ki povzroča COVID-19. izoliran od pacienta v ZDA, ki izhaja iz površine celic, gojenih v laboratoriju. Zasluga: NIAID-RML
* * *
Fotografija elektronskog mikroskopa Elizabeth Fischer prikazuje virusne čestice koje se oslobađaju iz umiruće stanice zaražene koronavirusom. Deseci malih, plavih sfera koje izlaze s površine bubrežne stanice same su čestice virusa. (Uz dopuštenje dr. Elizabeth Fischer / KHN / TNS)
*
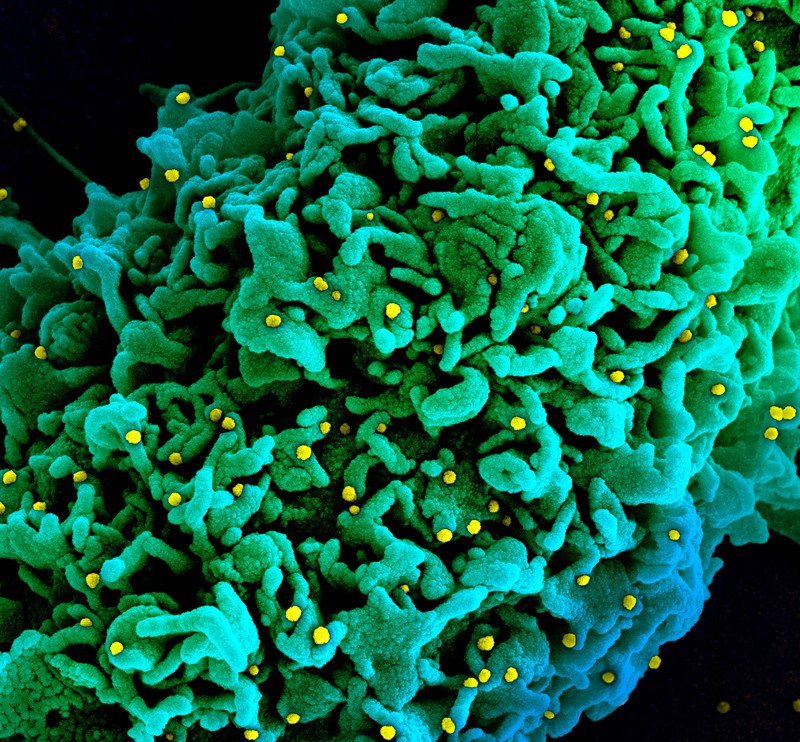
A cell infected with particles (yellow; artificially coloured) of the SARS-CoV-2 variant called B.1.1.7, which is more easily transmitted than other versions of the virus.Credit: National Institutes of Health/Science Photo Library
*
26.8. 2022: V mojih 45 letih raziskovanja in pisanja se nenehno pojavljajo miti, ki zamegljujejo ozadje. V večini primerov so bili ti miti del psihološke operacije (psyop), da bi odvrnili pozornost, razdelili ljudi in zmedli resnico. Na žalost preveč ljudi verjame laži in so nevtralizirani. Čas je, da končamo z mitom, da virus SARS-CoV-2 ne obstaja in da ni bil izoliran. Virus obstaja in je bil izoliran! ⁃ Urednik TN, Patrick Wood
dr. Meryl Nass: Še vedno me sprašujejo, ali je virus resničen. Na to vprašanje sem odgovorila v dveh prejšnjih objavah na blogu. Vendar je Off-Guardian objavil nov članek, v katerem trdi, da je virus ponarejen, zato so me ponovno prosili za komentar.
Ali je virus resničen? Ali je bil fotografiran? Kaj pa Kochovi postulati? Kaj storiti?
Neurejeno testiranje, pomanjkanje zanesljivih podatkov o Covidu-19 in hipoteza, zakaj
Tukaj je opis njegove kulture in izolacije, z dodatnimi podrobnostmi v dodatnem gradivu, za tiste, ki ga še naprej zahtevajo.
Izolacija in hitra delitev novega koronavirusa iz leta 2019 (SARS-CoV-2) pri prvem bolniku z diagnozo COVID-19 v Avstraliji
Ker sem v prejšnjih dveh objavah pojasnila nekatere tehnične vidike, bom kratka. Za več podrobnosti preberite komentar Johna McGowana k drugemu prispevku, ki pomaga ovreči članek Off-Guardian.
1. Prišlo je do izjemnega ponarejanja informacij o skoraj vseh vidikih pandemije. Mislim, da o tem ni veliko dvoma, in razumem, da so zaradi tega ljudje tudi primerno sumničavi glede resničnosti virusa. Še posebej, če ljudje z doktorati znanosti in doktorati znanosti za svojimi imeni trdijo, da virus ne obstaja.
2. Pripravljena sem zapisati, da se Andrew Kaufman, MD (psihiater, ne molekularni biolog, ki je diplomiral na istem oddelku kot jaz na MIT - biologija), ki je citiran v članku, moti in je poleg tega neveden. Tako kot drugi, ki zanikajo virus.
3. Tukaj je ključni argument: V izziv sem postavila tiste, ki zanikajo, da bi Covid povzročil pravi virus, naj pojasnijo, kaj točno povzroča te simptome, če ne gre za virus. Eden od predlaganih toksinov. Ali 5G.
Toksini, 5G in eksosomi niso nalezljivi, ta bolezen pa je.
Njena inkubacijska doba je zelo predvidljiva, v povprečju 6 dni.
Pravilno uporabljena osebna zaščitna oprema ščiti uporabnika pred izpostavljenostjo.
Pri tistih, ki zelo zbolijo, povzroča večinoma podobne sindrome.
Sindrom je sicer razmeroma edinstven, vendar je podoben tistemu, ki ga je leta 2003 povzročil SARS-1.
Ta preprosta dejstva, skupaj z argumenti, ki sem jih navedla v dveh povezanih prispevkih, potrjujejo, da se soočamo z nalezljivo boleznijo, ki jo povzroča virus.
4. Znanstveniki iz več držav so v mednarodne zbirke podatkov naložili na stotisoče zaporedij celotnega genoma (zemljevidi vsakega nukleotida v virusni RNA), pri čemer ima vsak od njih svoje lokalne mutacije. Te viruse bi morali izolirati in gojiti, da bi jih lahko sekvencirali. Če trdimo, da so vsa ta zaporedja napačna, moramo trditi, da na tisoče znanstvenikov skupaj laže o opravljenem delu. Ker so ti znanstveniki iz ZDA, Kitajske, Rusije in še od kod, jih ni enostavno pripraviti do tega, da bi vsi govorili isto laž.
5. Ali je ta virus nastal v laboratoriju? Skoraj zagotovo. Ali se je širil namerno? Ne vem. Lahko je bil naključen. Če je bil razširjen namerno, kdo je to storil? Tudi tega ne vem.
Če se tega vprašanja lotite z vprašanjem, kdo je imel sredstva, motiv in priložnost, da je storil tak zločin, lahko vsaj začnemo razpravo.
Bolezen se dobro odziva na protivirusna zdravila. Bolniki hitro ozdravijo, če se na začetku bolezni uporabijo protokoli za uničevanje virusov, vključno s hidroksiklorokinom ali ivermektinom.
a. Sredstva: Znanstveniki v več državah, vključno z ZDA in Kitajsko, so imeli sredstva za izdelavo takšnega virusa.
b. Motiv: Kdo ima koristi od pandemije? Za začetek ameriški dolar, Amazon, Elon, Facebook, Zoom, Twitter in država nadzora.
c. Priložnost: Ker so oktobra v Wuhanu potekale svetovne vojaške igre, je imelo vojaško osebje iz več deset držav priložnost. Wuhan je tudi mednarodno trgovsko središče. Morda je vsakdo, ki je lansko jesen obiskal Wuhan, imel priložnost.
Če je to teorijo tako enostavno ovreči, zakaj se še vedno pojavlja? Začenjam se spraševati, ali ne gre za psihološki poskus, ki ga vedno znova vnašajo v razpravo, da bi ljudem preprečili, da bi raziskali resnični izvor virusa... da bi raziskali financerje raziskav o koronavirusih na NIAID in drugod... in ugotovili, kaj natančno so poskušali narediti in za koga...
To so pomembna vprašanja, zlasti v smislu preprečevanja ponovitve pandemije, ki bi nastala v laboratoriju.
Prosim, ne zapravljajte več časa za hipotezo o "lažnem virusu". Nimamo časa ali razkošja, da bi se borili drug proti drugemu. Potrebujemo vse roke na krovu, da zavrnemo veliko ponastavitev (karkoli naj bi že bila) in si povrnemo zdrav nadzor nad našimi družbami.
*
If you want to map the tiniest parts of a protein, you only have a few options: You can coax millions of individual protein molecules to align into crystals and analyze them using x-ray crystallography. Or you can flash-freeze copies of the protein and bombard them with electrons, a lower resolution method called cryo–electron microscopy (cryo-EM). Now, for the first time, scientists have sharpened cryo-EM’s resolution to the atomic level, allowing them to pinpoint the positions of individual atoms in a variety of proteins at a resolution that rivals x-ray crystallography’s.
*
Odgovarja Dr. Alojz Ihan ...
Če prav razumem, želi kolega Gregor Knafelc prikaz/dokaz virusa SARS-CoV 2 direktno iz vzorca bolnika, pri tem pa »ne verjame« v virološke metode dokazovanja (PCR, kultivacija, sekvenciranje). Potem bi ga moral vprašati, v kaj od znanosti in stroke pa verjame? Če verjame v protitelesa in v imunohistokemijo, potem lahko v stotinah člankov vidi prikaz virusa v človeških tkivih, če le v zbirki PubMed vtipka »covid 19 immunohistochemistry« (Vir).
Če kolega verjame v elektronsko mikroskopijo, lahko virus v dihalih bolnikov prav tako občuduje v stotinah člankov (https://www.nature.com/articles/s41379-020-00661-1).
Če bi želel virus »ločiti od vsega ostalega«, ima na voljo kromatografijo ali imunoafinitetno kromatografijo, in ultracentrifugiranje (če »verjame« v te separacijske tehnike)- ampak potem mora po ločitvi »od vsega ostalega« še vedno verjeti vsaj v eno metodo dokazovanja virusa, na primer v imunofluorescenčno dokazovanje virusnih delcev, ali vsaj v encimsko imunske tehnike (ELISA), če je že PCR nekaj nepredstavljivega. Mimogrede – zakaj kolega ne verjame v PCR? Ali verjame v DNK in RNK? Če ja, potem morda ve, da gre za tako specifična zaporedja nukleotidov, da molekularni biologi hitro najdejo taka, ki so povsem unikatna za določen mikroorganizem in za nič drugega, vključno s človekom. Na tej tehniki danes temelji že tisoče rutinskih diagnostičnih testov, ne le mikrobioloških – kako potem kolega sploh verjame diagnozam, ki mu jih povedo pacienti?
*
Slika SARS CoV-2 posneta s krio-elektronsko tomografijo: https://www.sciencealert.com/this-technique-is-revealing-a-hidden-world-of-biology-weve-never-seen-before?utm_source=ScienceAlert+-+Daily+Email+Updates&utm_campaign=d71a26b84b-RSS_EMAIL_CAMPAIGN&utm_medium=email&utm_term=0_fe5632fb09-d71a26b84b-366127361
*
Lepo bi bilo torej, da bi kolega povedal, v katero od znanstvenih metod verjame, drugače mu bom težko razrešil njegov »etični problem«. Ker videti virus s prostim očesom ali ga otipati s prsti v nobenem primeru ne bo mogoče, tudi če mu ga kdo »loči od vsega ostalega«.
Morda pa kolegu čisto enostavno manjka nekaj dodatnega izobraževanja, ker je normalno, da človek po določenih letih od študija znanstveno nekoliko zarjavi. Namesto, da zahteva, naj se cel svet prilagodi njegovemu (ne)znanju, bi bilo pametneje, če bi se podobno kot Mohamed, raje sam odpravil h gori, ali vsaj na PubMed.
Za začetek pa povezave do učbenikov mikrobiologije in imunologije ter in infekcijskih bolezni, ki so pri nas izjemno sodobni in kvalitetni in so v njih odgovori na vse naštete dileme. V prvem od naštetih lahko na povezavi prebere celo (vzorčno) poglavje, v katerem je opis PCR in sekvenciranja, morda bo ob tem kolega celo našel nekaj svoje izgubljene »vere«:
Oktober 2021
*
Sodba španskega sodišča: virus ni izoliran. (op: ker je SARS CoV-2 virus patentiran in je biološko orožje): https://drcharlieward.com/breaking-news-spanish-high-court-confirms-that-covid-19-does-not-exist/?utm_content=13030304&utm_medium=Email&utm_name=Id&utm_source=Actionetics&utm_term=Email
*
14. tfSfapponroirlsmor sobS 1mloeef2lonfs:2r4rid · NEVERODOSTOJNO
Dr. Jerneja Tomšič ja zna in ima izkušnje. A žal je štartala iz napačne predpostavke. Zato je njena pripoved izgubila verodostojnoast. Virusi so bili odvzeti iz nosno žrelne votline pacienta s COVID 19. Virus je bil nacepljen na Vero-CCL81, v katerih se je bogato razmnoževal do visokih titrov. Izolacija datira v januar 2020. Tako namnožen virus je bil poslan tudi drugim laboratorijem. Torej virus je bil izoliran, je bil sekveniran je bil primerjan z genomom ostalih virusov, in izbran specifičen del genoma za potrjevanje v PCR testih.
* * *
Pa se to:
Prava izolacija naj bi bila precistiti virus od vsega ostalega, cesar nikoli niso naredili. Vedno je v supernatantu se drug genski material odmrlih celic in to sekvencirajo, da dobijo genom, ki je potem itak sestavljen s pomocjo racunalnika.
"Kdor se zadnji smeje, se najslajse smeje." - Jerneja Tomšič Caserta
Pomota.Preciscen v okužbi zivali ponovno izoliran v celoti po kochovih zakonih. Sekveniran primerjan s sorodnimi virusi. Pojma nimate. To ni za smejat ste za se zjokat. Kovid idi....
* * *
Odziv dekana UL Medicinske fakultete, prof. dr. Igorja Švaba dr. med., na objavo v medijih
V preteklem tednu se je na spletu pojavil posnetek, v katerem je gibanje Osveščeni prebivalci Slovenije za predstavitev svojih idej zlorabilo ime dekana UL Medicinske fakultete (UL MF), prof. dr. Igorja Švaba, sklicujoč se na odločbo UL MF dne 30. novembra 2020:Ta dokument, ki se pojavlja na posnetku, ni nikakršen dokaz, da virus ne obstaja.
Gre za dokument, v katerem UL MF trdi, da UL MF ni izvajala znanstvene študije "o izolaciji virusa Sars-Cov-2 v skladu s Kochovimi postulati", ki jo je specificiralo vprašanje. Kochovi postulati, objavljeni leta 1890, so bili narejeni za dokaz bakterijskih okužb, viruse so odkrili kasneje in tudi metode njihovega dokazovanja so povsem drugačne. Kochovi postulati na primer zahtevajo gojenje povzročitelja na sterilnem gojišču, viruse pa je mogoče gojiti samo na celičnih kulturah. Ker torej vprašanje, naslovljeno na UL MF, sprašuje po strokovno nesmiselnih dokazih, je UL MF odgovorila, da s takimi dokazi ne razpolaga.Stališče UL MF je, da je obstoj virusa Sars-Cov-2 nedvomno dokazan s sodobnimi metodami, s katerimi se dokazuje obstoj, prenosljivost in patogenost virusov.UL MF dosledno zagovarja znanstveno utemeljena stališča glede covid-19 in da podpira in spoštuje ukrepe za omejevanje te bolezni, vključno s cepljenjem in pogoji PCT.Dekan UL Medicinske fakultete, prof. dr. Igor Švab, ni avtor nobenega izmed dokumentov, ki zanikajo obstoj virusa SARS-Cov-2 in sporoča, da gre po njegovem mnenju pri objavi gibanja Osveščeni prebivalci Slovenije za predstavitev svojih idej za nedopustno zlorabo imena in položaja medicinskega strokovnjaka. UL Medicinska fakulteta sprejema in resno jemlje svoje poslanstvo ter sodeluje pri znanstveno podkrepljenem obveščanju laične in strokovne javnosti o vseh bolezenskih stanjih, ne samo covid-19. V preteklih mesecih smo tako organizirali številne javnosti dostopne seminarje, na katerih so znanstveniki odgovarjali na odprta vprašanja ter pojasnjevali široko področje bolezni covid-19. Vsa gradiva so še vedno dostopna javnosti in priporočamo ogled vseh vsebin: https://lifelong.mf.uni-lj.si/course/index.php?categoryid=29
*
Volume 26, Number 9—September 2020
Research
Isolation, Sequence, Infectivity, and Replication Kinetics of Severe Acute Respiratory Syndrome Coronavirus 2
Arinjay Banerjee, Jalees A. Nasir1, Patrick Budylowski1, Lily Yip, Patryk Aftanas, Natasha Christie, Ayoob Ghalami, Kaushal Baid, Amogelang R. Raphenya, Jeremy A. Hirota, Matthew S. Miller, Allison J. McGeer, Mario Ostrowski, Robert A. Kozak, Andrew G. McArthur, Karen Mossman , and Samira Mubareka
Author affiliations: McMaster University, Hamilton, Ontario, Canada (A. Banerjee, J.A. Nasir, K. Baid, A.R. Raphenya, J.A. Hirota, M.S. Miller, A.G. McArthur, K. Mossman); University of Toronto, Toronto, Ontario, Canada (P. Budylowski, N. Christie, A. Ghalami, A.J. McGeer, M. Ostrowski, R.A. Kozak, S. Mubareka); Sunnybrook Research Institute, Toronto (L. Yip, P. Aftanas, R.A. Kozak, S. Mubareka); Mount Sinai Hospital, Toronto (A.J. McGeer)
Vir: https://wwwnc.cdc.gov/eid/article/26/9/20-1495-f1
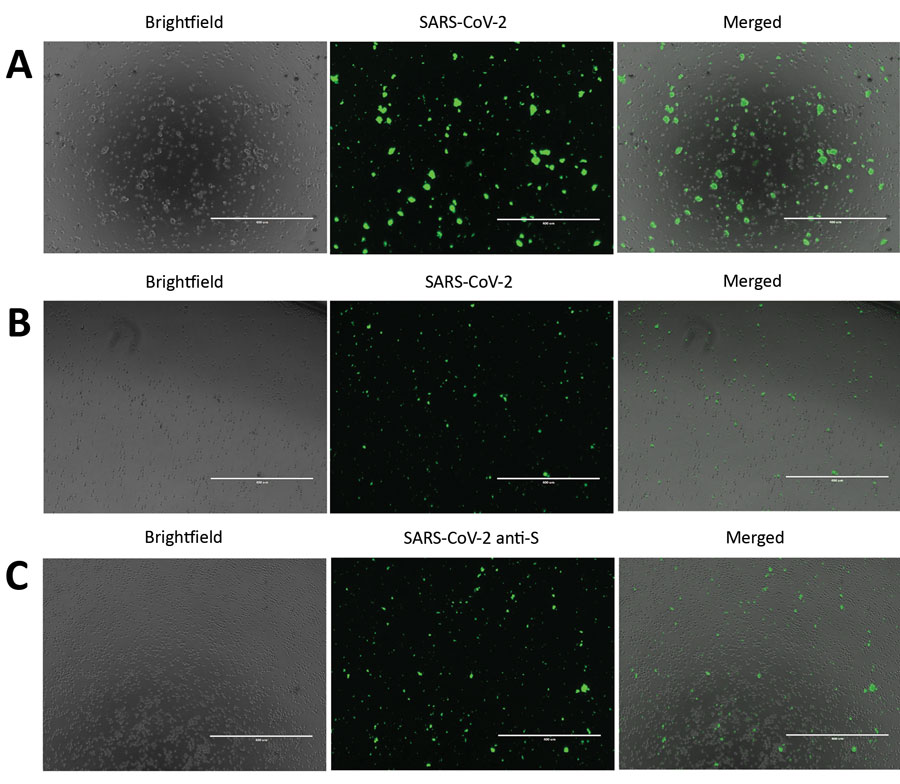
Slika 1. Odkrivanje hudega akutnega respiratornega sindroma koronavirusa 2 (SARS-CoV-2) v okuženih celicah Vero E6 in CD4 + T. Da bi odkrili izražanje beljakovin SARS-CoV-2, smo okužili celice Vero E6 in CD4 + T z množico virusov SARS-CoV-2 za 24 ur. Te celice smo imunsko obarvali in jih opazovali s fluorescenčno mikroskopijo. A) SARS-CoV-2 so okužili in imunsko obarvali celice Vero E6. B) SARS-CoV-2 virusi so okužili in imunsko obarvali celice CD4 + T. Za plošči A in B so celice obarvali z uporabo protitelesnega koktajla, sestavljenega iz protiteles SARS-CoV-2 S1, protiteles SARS-CoV-2 N in razredčenega seruma bolnika, ki je prebolel koronavirus. C) CD4 + T celice, okužene s SARS-CoV-2, imunsko obarvane s protitelesi SARS-CoV-2 S1 (anti-S). Merilne črte označujejo 400 μm; prvotna povečava × 10.
Povzetek
Od njegovega pojava v Wuhanu na Kitajskem decembra 2019 je hud akutni respiratorni sindrom koronavirus 2 (SARS-CoV-2) okužil ≈6 milijonov ljudi po vsem svetu. Ko se SARS-CoV-2 širi po vsem planetu, smo raziskali vrsto človeških celic, ki jih lahko okuži ta virus. Izolirali smo SARS-CoV-2 od dveh okuženih bolnikov v Torontu v Kanadi; določili genomska zaporedja; in ugotovili enonukleotidne spremembe v reprezentativnih populacijah naših zalog virusov. Preizkusili smo tudi širok spekter človeških imunskih celic za produktivno okužbo s SARS-CoV-2. Potrjujemo, da človeške mononuklearne celice periferne krvi ne dovoljujejo SARS-CoV-2. Ker se SARS-CoV-2 še naprej širi po vsem svetu, je bistvenega pomena spremljanje enonukleotidnih polimorfizmov virusa in nadaljevanje izolacije virusov v obtoku za določanje virusnega genotipa in fenotipa z uporabo modelov okužbe "in vitro" in "in vivo".
Ko se SARS-CoV-2 širi po vsem svetu, se virus verjetno prilagaja in razvija. Nujno je izolirati viruse SARS-CoV-2, da označimo njihovo sposobnost okužbe in razmnoževanja v več tipih človeških celic ter ugotovimo, ali se virus razvija v svoji sposobnosti, da okuži človeške celice in povzroči hudo bolezen. Izolacija virusa ponuja tudi priložnost, da virus deli z drugimi raziskovalci za razvoj in testiranje diagnostike, zdravil in cepiv.
Iz dveh bolnikov s COVID-19 smo izolirali SARS-CoV-2 in določili genomsko zaporedje vsakega izolata (SARS-CoV-2 / SB2 in SARS-CoV-2 / SB3-TYAGNC). Poleg tega smo preučevali replikacijsko kinetiko SARS-CoV-2 / SB3-TYAGNC v človeških fibroblastih, epitelijskih in imunskih celicah.
Razprava
Poročali smo o izolaciji dveh replikacijskih kompetentnih vzorcev virusa SARS-CoV-2 od bolnikov s COVID-19 v Kanadi. Za olajšanje izolacije virusa iz kliničnih vzorcev smo uporabili s TPCK obdelani tripsin (slika 2, plošča A). Eksogeni tripsin učinkoviteje aktivira beljakovine bodic (spike) SARS-CoV in olajša vstop v celice (21). Z obravnavo z eksogenoim tripsinom se poveča tudi nalezljivost drugih zoonotičnih netopirjevih koronavirusov (22). Poleg tega so s TPCK-tretiranim tripsinom na Kitajskem uspešno izolirali SARS-CoV-2 (1). V naši študiji za poznejšo okužbo in razmnoževanje virusa ni bil potreben noben dodaten tripsin, tretiran s TPCK (slika 2, plošča B). Prisotnost samega CPE ne pomeni uspešne izolacije koronavirusa. Vzorci srednje turbinate pri odraslih z akutno dihalno stisko lahko pogosto vsebujejo druge mikrobe, vključno z virusi (23). Za identifikacijo izolatov naše celične kulture smo jih sekvencirali, da potrdimo, da odražajo okužbe bolnikov s SARS-CoV-2 po vsem svetu, in za eksperimentalno preiskavo izbrali SARS-CoV-2 / SB3-TYAGNC, ker je ta izolat povzročil manj odčitkov zaporedij.
SARS-CoV je povzročil izbruh hudega akutnega respiratornega sindroma v letih 2003–2004. SARS-CoV lahko okuži človeške strukturne (24) in imunske celične linije (25) "in vitro". Da bi prepoznali vrste celic, ki lahko podpirajo produktivno okužbo s SARS-CoV-2, smo s SARS-CoV-2 / SB3-TYAGNC okužili vrsto populacij človeških celic. Tako celice Vero E6 kot Calu-3 podpirajo replikacijo SARS-CoV-2 na visoke titre (slika 3), kot poročajo druge nedavne študije (26,27). Prej je bilo tudi dokazano, da se SARS-CoV učinkovito replicira v celicah Vero E6 (24). Celice Vero E6 so imunsko pomanjkljive in imajo pomanjkljivo prirojeno signalizacijo protivirusnega interferona, zaradi česar so idealni kandidati za izolacijo virusa (28). Da pa omogočimo študije o interakcijah SARS-CoV-2 in gostitelja, je pomembno prepoznati človeške pljučne epitelijske celice s pravilnimi imunskimi odzivi, ki lahko podpirajo replikacijo SARS-CoV-2. Mi in drugi smo že pokazali, da se korona virus SARS-CoV in bližnjevzhodni respiratorni sindrom (MERS-CoV) učinkovito replicirata v celicah Calu-3 (8,29,30). Poleg tega so v celicah Calu-3 preučevali imunski odziv, ki ga povzroča SARS-CoV in MERS-CoV (30,31). Sposobnost okužbe celic Calu-3 s SARS-CoV-2 (slika 3) bo olajšala in vitro študije interakcij virus-gostitelj z uporabo SARS-CoV-2. Druge pogosto uporabljene človeške pljučne celice, kot je A549, ne podpirajo učinkovite replikacije SARS-CoV-2 (26). Poleg tega celice THF hTERT (človeška telomeraza reverzna transkriptaza) prav tako niso podpirale replikacije virusa (slika 3).
Prejšnje študije so pokazale, da so človeške imunske celice, kot so celice THP-1, dovzetne za okužbo s SARS-CoV (25). V naši študiji populacije človeških imunskih celic, vključno s celičnimi linijami, pridobljenimi iz THP-1, in primarnimi celicami (PBMC) niso podpirale produktivne replikacije SARS-CoV-2 (slika 3). Čeprav primarne CD4 + T celice niso podpirale produktivne replikacije virusa, smo z elektronsko mikroskopijo v teh celicah opazili virusom podobne delce (slika 4, plošča C). Tudi s pomočjo fluorescentne mikroskopije smo zaznali proteine SARS-CoV-2 v okuženih CD4 + T celicah (slika 1, plošči B, C). Ta ugotovitev je v skladu s tisto, o kateri so nedavno poročali Wang in sod. ko so dokazali, da lahko virusi SARS-CoV-2 in psevdotipizirani virusi vstopijo v človeške T-celične linije (MT-2) (32). Ti avtorji so tudi ugotovili, da je bila replikacija SARS-CoV-2 v celicah MT-2 neučinkovita. Raven transkripcije SARS-CoV-2 v okuženih celicah MT-2 se je povečala po 6 urah po okužbi, vendar je ostala stabilna tudi pozneje, kar kaže na pomanjkanje replikacije virusa v teh celicah (32). Ta ugotovitev je podobna neuspešni replikaciji, opaženi pri limfocitih T, okuženih z MERS-CoV (33). Vendar pa študija Wang in sod. ni količinsko opredelila titrov virusa v supernatantu iz okuženih celic. V naši študiji v supernatantu, ki je bil zbran iz CD4 + T celic, okuženih s SARS-CoV-2, nismo mogli zaznati nobenega virusa, ki bi bil sposoben za replikacijo (slika 3). Človeške imunske celice nimajo izražanja encima, ki pretvarja angiotenzin 2 (34) (https: //www.proteinatlas.orgExternal Link), ki je funkcionalni receptor za SARS-CoV-2 (1,35). Novi podatki kažejo, da bi lahko obstajali tudi drugi receptorji, kot je CD147, ki bi lahko olajšali celični vnos SARS-CoV-2 (K. Wang et al., Neobjavljeni podatki, https://www.biorxiv.org/content/10.1101 /2020.03.14.988345v1Zunanja povezava). Potrebne so dodatne študije za določitev celotnega razpona celičnih receptorjev in koreceptorjev, ki lahko olajšajo vstop SARS-CoV-2. Torej, čeprav je zanimivo, da so celice CD4 + T lahko dovzetne za SARS-CoV-2, naši podatki kažejo, da te celice "in vitro" ne dopuščajo replikacije SARS-CoV-2. Potrebnih je več študij, da bi v celoti ugotovili učinke vnosa SARS-CoV-2 v CD4 + T limfocite.
V zaključku poročamo, da čeprav človeška pljučna celična linija podpira replikacijo SARS-CoV-2, se virus ni razmnožil v nobeni od preizkušenih imunskih celičnih linij ali primarnih človeških imunskih celicah. Čeprav v primarnih limfocitih CD4 + nismo opazili produktivne okužbe, smo v teh celicah z elektronsko mikroskopijo opazili virusom podobne delce. Tako lahko SARS-CoV-2 vstopi v CD4 + primarne limfocite T, vendar se ne more učinkovito razmnoževati. Naši podatki osvetljujejo širši spekter človeških celic, ki lahko ali pa ne omogočajo razmnoževanja SARS-CoV-2, naša študija pa močno nakazuje, da testirane človeške imunske celice ne podpirajo produktivne okužbe s SARS-CoV-2
Vir: https://wwwnc.cdc.gov/eid/article/26/9/20-1495_article
https://www.medrxiv.org/content/10.1101/2020.08.01.20166553v2
*
Neposreden dokaz SARS CoV-2 koronavirusa v črevesnem endoteliju
11. september 2020
Klaus Stahl,
Jan Hinrich Bräsen,
Marius M. Hoeper &
Sascha David
Intensive Care Medicine volume , pages
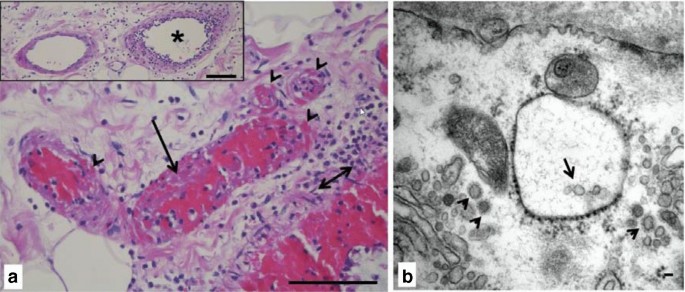
Klinično opažanje, da lahko koronavirusna bolezen (COVID) -19 prizadene različne organe, je bilo nedavno pripisano okužbi endotelijskih celic s SARS-CoV-2, ki vodi do endotelialitisa in mikrovaskularne koagulacije. To znanje razširimo s prikazom patoloških ugotovitev iz vzorca črevesja. Prej zdrav, 43-letni moški je po stiku s SARS-CoV-2 razvil gripi podobne simptome. Dva tedna kasneje je imel odpoved dihanja, ki je zahtevala endotrahealno intubacijo, mehansko ventilacijo in zunajtelesno membransko oksigenacijo. Klinični potek se je podaljšal s počasnim okrevanjem pljučne funkcije in stalnimi znaki sistemskega vnetja. Štiri tedne po intubaciji se je pojavila akutna neobstruktivna mezenterična ishemija in potrebna je bila nujna hemikolektomija. Histološki pregled je razkril hud endotelialitis in več mikrotrombov, zlasti v venski žilni postelji (puščice na sliki 1a).
Elektronska mikroskopija je pokazala več virusnih delcev SARS-CoV-2 v endoteliju debelega črevesa (slika 1b), ki je znano po nadpovprečni izraženosti vstopnega receptorja ACE-2. Omeniti velja, da je neobstruktivna nekroza črevesja vse pogosteje prepoznana kot nov zaplet hude bolezni COVID-19. Preseneča nas, da smo v endoteliju črevesja našli približno in na videz nedotaknjene virusne delce približno 8 tednov po začetni okužbi, ko virusa že nismo mogli zaznati v vzorcih dihal in krvi.
a Submukozne žile na vzorcu hemikolektomije kažejo hud endotelialitis, pretežno v venskih žilah (vstavljena zvezdica) z nastankom trombov (dolga puščica) in krvavitvijo v žilno steno (konice puščic). Madež H&E, dvojna puščica označuje sluznico lamina muscularis, stolpci predstavljajo 100 µm. b Elektronska mikroskopija venske endotelijske celice razkrije koronaviruse (majhne puščice) in vezikle, ki vsebujejo delce viriona (puščica). Vzorec vgrajenega epona v kontrastni taninski kislini, stolpec predstavlja 50 nm
Vir: https://link.springer.com/article/10.1007/s00134-020-06237-6
*
Neuporabnost PCR testa https://off-guardian.org/2020/11/17/covid19-evidence-of-global-fraud/
*
Podobnost med nekaterimi genskimi zaporedji SARS CoV-2 in človeškega genoma
https://virological.org/t/response-to-ncov2019-against-backdrop-of-endogenous-retroviruses/396
*
dec 2020: The "new" British variant B.1.1.7 has a mutated ORF8 gene. These mutations have been seen previously, and were less virulent/deadly in Africa, France, and Singapore. People who catch it have never required supplemental oxygen.
https://www.medrxiv.org/content/10.1101/2020.12.23.20248758v1
*
Virusi MERS EMC 2012 na površini celice- beta koronavirus bližnje vzhodnega respiratornega sindroma MERS, ki je povzročil kameljo gripo leta 2012
*
Sekvencioniranje SARS CoV-2 na Nizozemskem
SARS CoV-2 vsebuje 29.902 nukleotidov (črk)
Vir: dr. Gorazd Pretnar 20. 12. 2020 FB
*
Sekevencionirnje virusov: https://www.illumina.com/areas-of-interest/microbiology/infectious-diseases/coronavirus-sequencing.html?fbclid=IwAR17vAYy_3FsN-MfL4ypwFdgfcBKaMSwY25dkSVv2JKyqJy0DBlRC_kgUO8
*
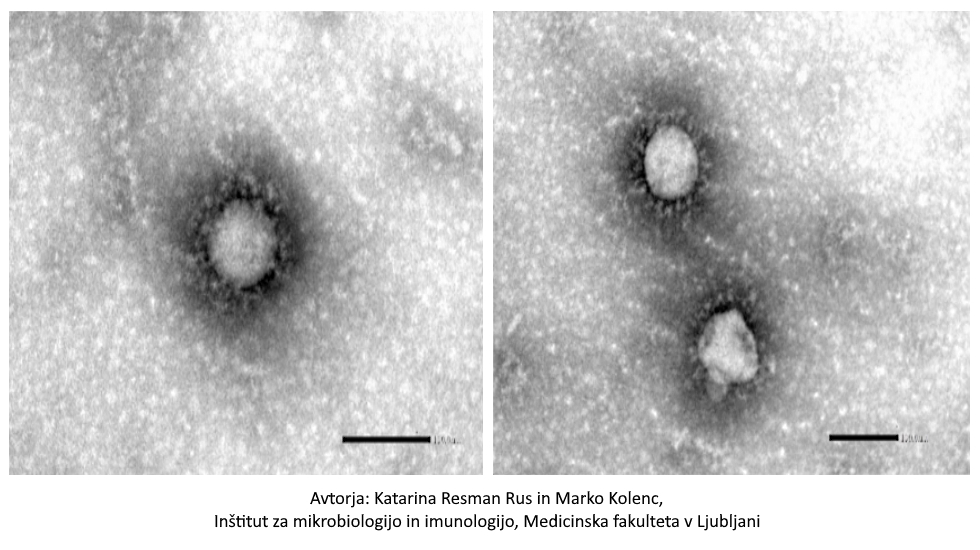
ztok Konc 14. 01. 2021
Fotografija »slovenskega« koronavirusa SARS-CoV-2
Na vprašanja in dvome o covidu odgovarja najbolj ugledna slovenska virologinja akad. Tatjana Avšič Županc
Vir: https://prvi.rtvslo.si/podkast/ultrazvok/2288722/174746073
V množici zapisov, prispevkov in člankov, še zlasti na spletu in družbenih omrežjih, nekateri še vedno dvomijo v resničnost in točnost informacij o covidu. Ali je fotografija lahko dokaz, da novi koronavirus obstaja? Kaj pa dejstvo, da poznamo njegov genski zapis? Kaj se zgodi z virusom, ko covid prebolimo? Zakaj PCR test večkrat ponovijo? Kako učinkovito je cepivo?
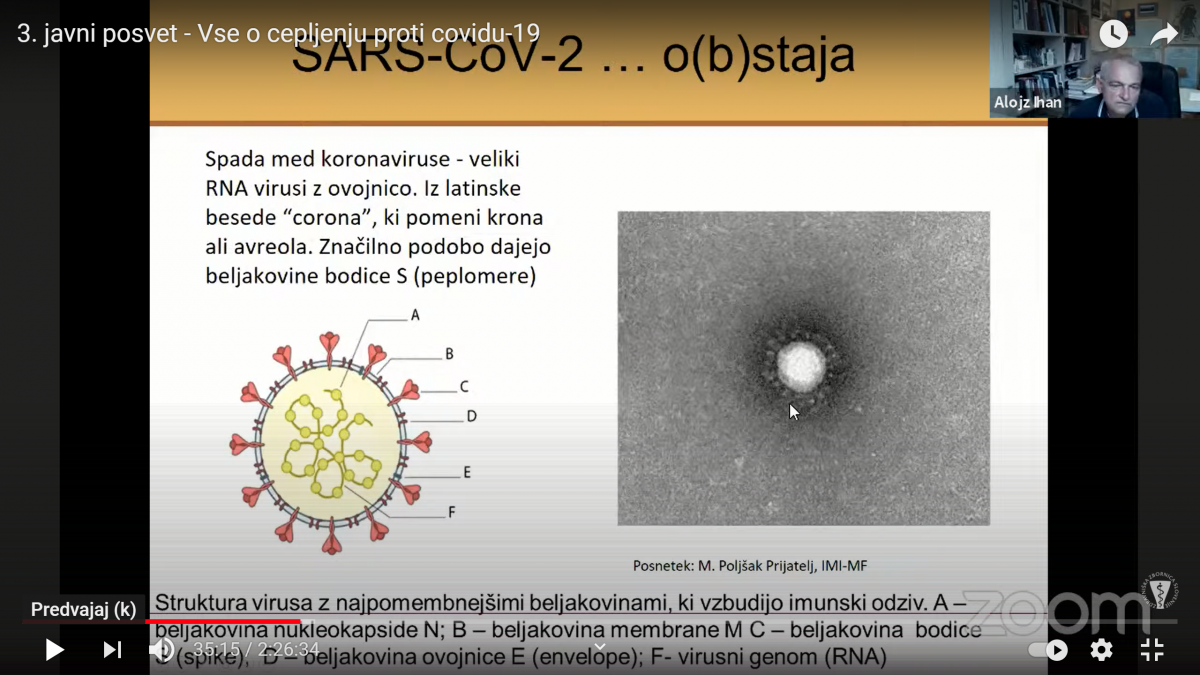
Opis fotografije: Virus SARS-CoV-2, posneta s transmisijskim elektronskim mikroskopom JEOL TEM-1400 Plus; negativno kontrastiranje 2% FWK; povečava 100 000x in 80 000x.
V Ultrazvoku tokrat na vprašanja o novem koronavirusu in covidu odgovarja najbolj ugledna slovenska virologinja akad. prof. dr. Tatjana Avšič Županc z Inštituta za mikrobiologijo in imunologijo Medicinske fakultete v Ljubljani.
*
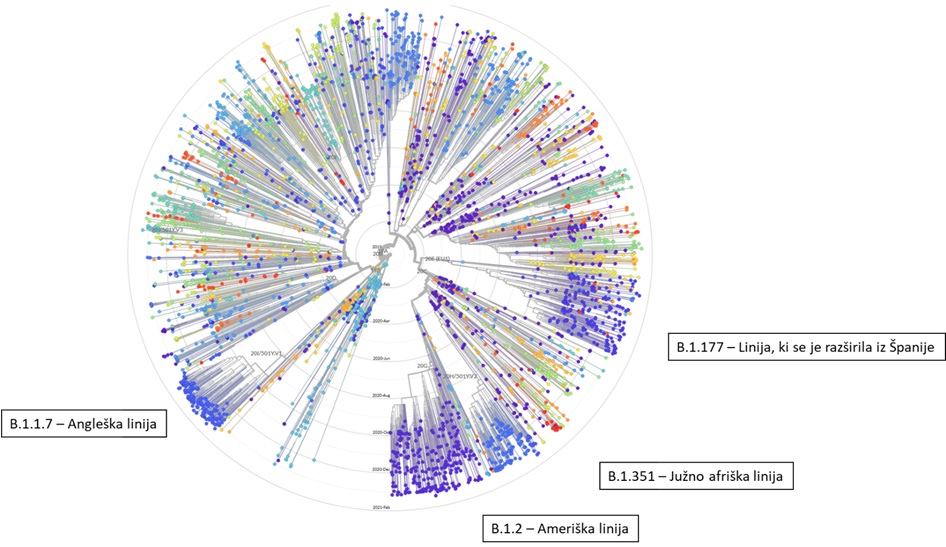
Razvoj in mutacije koronavirusa SARS CoV-2
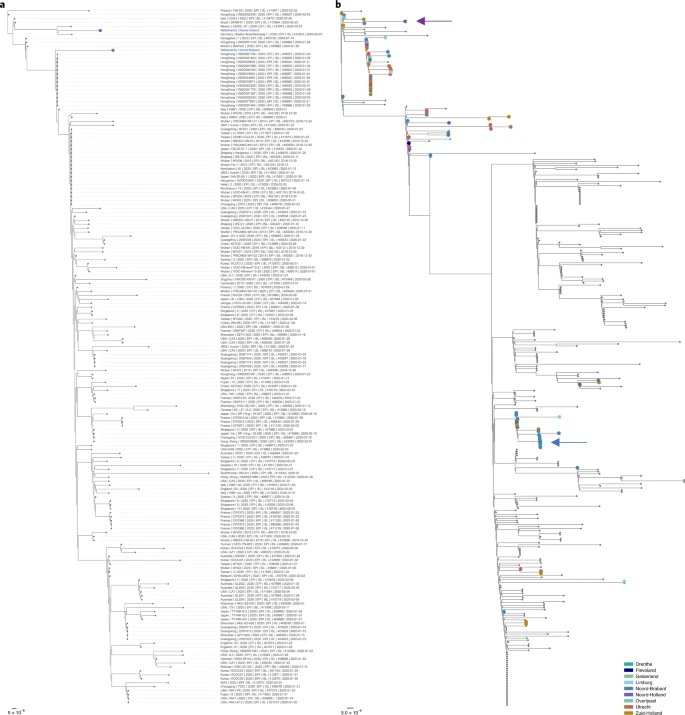
The analysis shows there are currently seven main strains of the virus. The original strain, detected in the Chinese city of Wuhan in December 2019, is the L strain. The virus then mutated into the S strain at the beginning of 2020. That was followed by V and G strains. Strain G mutated yet further into strains GR, GH and GV. Several other infrequent mutations were collectively grouped together as strain O.
The graphic above shows how the original L strain is almost gone, leaving G strains dominant in the current stage of the pandemic. That’s important because the G strains include one mutation that makes it easier for the spike proteins on SARS-CoV-2 to bind to receptors on human cells, potentially increasing the chances of infection and transmissibility of the virus.
*
Celotni genom SARS CoV-2 izoliran v Nemčiji
https://mra.asm.org/content/9/23/e00520-20.abstract
*
*
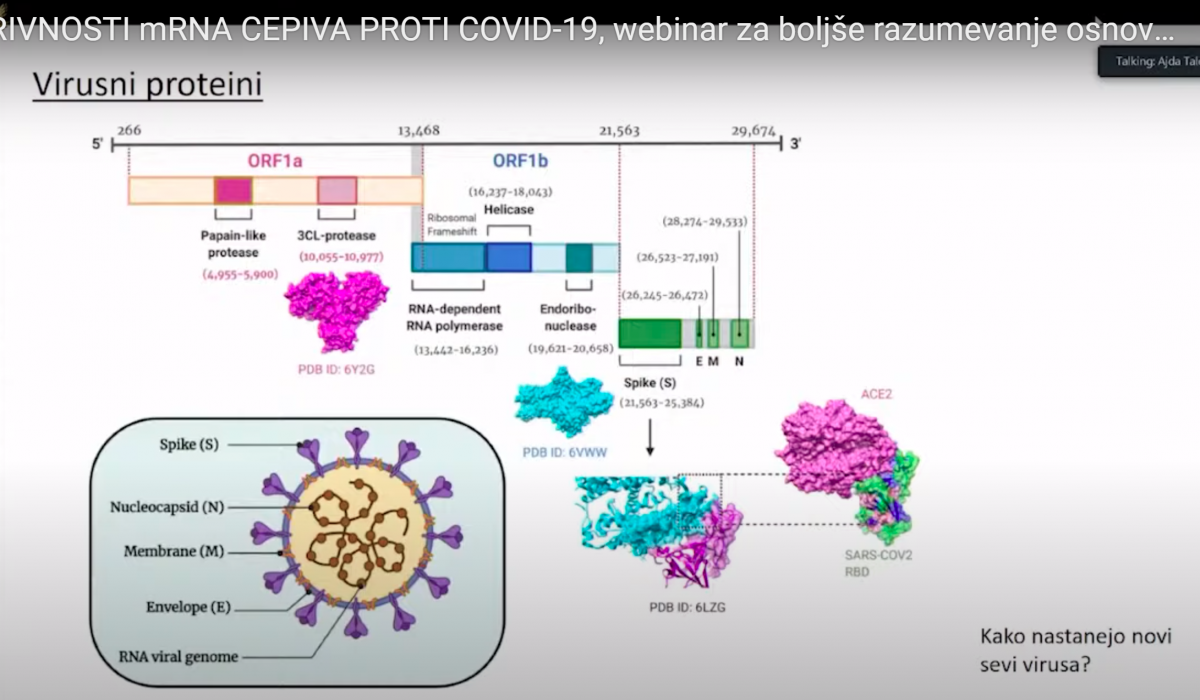
Virusni proteini- predavanje dr. Ajda Taler Verčič 28.1. 2021
S protein(beljkovinska bodica) je kodiran z nukleotidi od 21.563-26.472 v genomskem zaporedju
*
Virusni dedni material si namreč lahko predstavljamo kot 30.000 črk dolgo verižico, na kateri so v točno določenem vrstnem redu nanizane črke A, U, G ali C. Pri nastajanju novih verižic, to je novih genomov, se posamezne črke v novem virusnem genomu pomotoma in naključno zamenjajo. Tako iz enega ‘starša’ nastanejo različni ‘potomci’, kar lahko predstavimo podobno kot družinsko drevo. Večina teh sprememb ne vpliva na obnašanje virusa, zato so virusi enakomerno zastopani in vse veje v drevesu so bolj ali manj enako debele. Občasno pa se pojavi verzija virusa, ki se zelo dobro razširja in zato postane posamezna veja bolj opazna. Take variante potem poimenujemo linija ali pa sev, recimo angleški sev ali južnoafriški sev. To poimenovanje je zelo poljudno in ni znanstveno. Pravilneje je uporabiti izraz ‘različica’ (varianta).
*
https://www.cdc.gov/coronavirus/2019-ncov/lab/grows-virus-cell-culture.html
SARS-CoV-2, the virus that causes COVID-19, was isolated in the laboratory and is available for research by the scientific and medical community.
One important way that CDC has supported global efforts to study and learn about SARS-CoV-2 in the laboratory was by growing the virus in cell culture and ensuring that it was widely available. Researchers in the scientific and medical community can use virus obtained from this work in their studies.
SARS-CoV-2 strains supplied by CDC and other researchers can be requested, free, from the Biodefense and Emerging Infections Research (BEI) Resources Repositoryexternal icon by established institutions that meet BEI requirements. These requirements include maintaining appropriate facilities and safety programs, as well as having the appropriate expertise. BEI supplies organisms and reagents to the broader community of microbiology and infectious disease researchers.
Poročilo obdukcij v UK: https://www.sciencedirect.com/science/article/pii/S1756231720302292?fbclid=IwAR3wqqOJmJewwH1d52z2WlAej12kq6J3y0AX4PvbnxJBZLnF7XZRn8aPL5Y
*
Unievrza LJ priznala da nimajo virusa izoliranega in dokaza, da povzroca covid 19
14.4. 2021
Zakaj bi ga po vase morali na mf izolirat. Prosim odgovorite mi razumno in logicno
Morali bi izolirat, da bi imeli izoliran virus kot pozitivno kontrolo pri pCR testu naprimer. Pa nihce na mf LJ tudi ni dokazal, da so simptomi povzroceni od virusa in to niso dokazali niti drugje.
Pozitivno kontrolo se dobi v banki podarjeno ali kupljeno. Nekaj ponavljat kar je veckrat ponovljeno nima smisla. Sam kupujem pozitivne kontrole bakterij v preverjenih bankah. Prosim nehajte klatit neumnosti. V znanosti ni mej sodelujemo drug z drugim.Se posebej sedaj. In nehajte pljuvat po znanosti ce niste del nje. Obrnes tel dobis ali das. Si ne predstavljate. Trditev da virus ni mogoce izolirat je bedarija. Ce to trdis ne gre z jezikom kot gospa ampak s poizkusi. Ste naredil neskoncno skode z neznanjem.
Pa drži informacija, da je treba pridobiti virus iz prvega bolnika?
Tunga Vidya Dasi
Seveda drži. Teh izolacij je bilo v svetu veliko, in razmnožitvi na celicah tudi.
Ampak jaz sem nekje prebrala, da Kitajci tega niso dali.
Tunga Vidya Dasi
One hundred microliter of oropharyngeal and nasopharyngeal swab specimen of these cases was inoculated onto 24-well cell culture monolayers of Vero CCL-81 and incubated for an hour at 37°C to allow virus adsorption, with rocking every 10 minuts.
https://www.youtube.com/watch?v=hqe3MhV9-MU
Gorazd Janez Pretnar
Ti ljudje, ki vam očitajo, da niste izolirali virusa, dobro vejo za te študije, ki jih tukaj delite, ampak pravijo, da niso zadostne, ker ne gre za pravo izolacijo virusa, ampak za množenje na celicah v prisotnosti strupenih kemikalij, ki v celicah sprožijo izločanje "virusov" (v resnici eksosomov, ki jih tvori sama celica pod stresom), ki jih virologi potem analizirajo. A resno mislite, da Jerneja Tomšič Caserta še ni videla študij, ki ste jih delili o izolaciji?
Tukaj imate vse skupaj lepo opisano: https://theinfectiousmyth.com/.../IsolationVersusPurifica...
1500 testov ni bilo pozitivnih na SARS CoV-2, temveč na virus influenza A in B
Pika Dobrovoljec
Bedarija: moj kolega je to raztrančiral v tem prispevku.
https://www.youtube.com/watch?v=_vHx1x0TCAY
https://science.sciencemag.org/content/370/6513/203
Naj sama pove. Vi pa kaj preberite o eksosomih. In ce mislite da so nasi labi dejansko onesnazeni hlevi vam ni pmoci. Iscete neskoncno bedastih argumentov za diskreditacijo znanosti. Molekularka naj mi pove koliko in katere viruse je v zivljenju izolirala. In katere celicne kulture uporablja in v kaksnih pogojih.
Gorazd Janez Pretnar
Zelo slabi argumenti so tole! Tale vaš kolega omenja npr. študije na miših, ki naj bi dokazovale patogenost "virusa". Tole je v enem od znanstvenih člankov napisano o teh študijah:
“Although useful, [hACE2 transgenic mice models] produced variable results in which some supported low level SARS-CoV-2 replication with minimal morbidity or lung pathology [22], and others exhibited signs of local and systemic disease which partially mimic symptoms of severe human COVID-19 infection, including increased mortality in male mice [23].”
Vir:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7655046/...
Kako to, da so nekatere od študij na teh miših ugotavljale, da virus sploh ni blazno nevaren in povzroča le minimalno okuženost in patologijo? Če je virus res hudo nevaren, bi moral biti v vseh študijah hudo nevaren, pa ni, torej je nekaj zelo narobe s študijami. Spet druge študije ugotavljajo povišano smrtnost le pri mišjih samcih. Kje je logika?
Tale članek lepo opiše vse nesmisle, ki jih producirate virologi: https://drtomcowan.com/what-does-a-virologist-know-about.../
Med drugim piše, da "virus" razmnožuje le na celicah opičjih ledvic (Vero) in praktično na nobenih drugih celicah. Kje je tukaj logika? Hudo nevaren virus, ki prizadene vse organe, pa se ne razmnožuje na praktično nobenih celicah??
https://drtomcowan.com/what-does-a-virologist-know-about-sars-cov2/?fbclid=IwAR36Hf97quhVaLVSy2qYCYo9R2sL3mNKhe7cIFIKJlCj66N8btwodAHO3TY
Kar se tiče eksosomov pa izgleda, da vam manjka znanja o njih. Predlagam, da začnete s tole študijo: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7291340/
"In recent decades, the similarity between EVs and viral particles has become increasingly evident. Viruses and EVs share different aspects such as size, structural and biochemical composition, and the transport of bioactive molecules within cells [34,35]. Like EVs, viruses present a size ranging from 30 to 1000 nm, starting from the small ones, such as poliovirus and hepatitis A virus (HAV) particles, which possess a diameter of about 30 nm, all the way to hepatitis C virus (HCV) of about 50 nm, and HIV or SARS viruses that are about 100–120 nm. Finally, mimiviruses have a size of about 400 nm. Furthermore, EVs and some viruses have morphological similarities: as previously described, EVs are double-membrane-enclosed entities and enveloped viruses are also surrounded by a lipid membrane acquired from the cell. Interestingly, they possess a similar lipid composition enriched in glycosphingolipids and cholesterol, as well as a similar protein content. Notably, both EVs and viruses carry nucleic acids; while viruses present single or double-stranded RNA or DNA genomes, which are carried and protected inside their capsid, EVs can transport a variety of nucleic acids [35,37,38]. EVs and enveloped viruses also share similar biogenesis processes since both are generated in the endosomal network or bud from the plasma membrane using specific pathways [18]. For example, some retroviruses such as HIV hijack the cellular vesiculation machinery to favor their own replication and budding. In this regard, it has been reported that the endosomal sorting complex (ESCRT), the same that mediates the inward invagination of ILVs in MVBs, is also involved in the budding and release of HIV particles [39,40]. Moreover, just as EVs can be generated from ESCRT-independent pathways, some viruses bud from specific membrane domains [41]. These domains, called lipid rafts, are enriched in glycosphingolipids, cholesterol and ceramide. In addition, proteins like tetraspanins are stored in these domains and form clusters among themselves and other transmembrane and cytosolic proteins, thus inducing inward budding of the microdomains in which they are enriched [42]. As previously mentioned, specific glycocalyx compositions also play a role in vesicle release; however, glycocalyx can be also involved in other membrane processes, including the absorption of some viruses [43]. In this regard, some viruses have evolved to exploit specific glycans to enter cells, like human rotaviruses that bind the blood group A antigens [44]. Instead, in the case of HIV [45], Ebola virus [46], HCV [47], as well as influenza [48] or Severe Acute Respiratory Syndrome (SARS) viruses [49], the viruses themselves present glycans on their surface. Their presence on viral surfaces is exploited by immune cells, such as macrophages or dendritic cells, to phagocyte virions. In turn, Ebola [46] and SARS viruses [49] take advantage of this anti-viral system to enter and replicate in macrophages and dendritic cells. On the other hand, glycans are also used by viruses to create a shield that hides viral epitopes to immune cells, as happens with HIV, known to have the highest density of glycans attached to its surface proteins [50], and the Lassa virus [51].
The substantial overlap of the biogenesis processes provides a plausible explanation for the similar composition observed between EVs and enveloped viruses [39]. Furthermore, both EVs and enveloped viruses can bind to the plasma membrane of recipient cells and, after fusion events, directly with the surface membrane or after endocytosis, they release their luminal cargo into the cytosol, influencing cell activity [18]. In this respect, in a similar manner to the viral envelope proteins, EV surface proteins, such as the intercellular adhesion molecule 1 (ICAM-1), mediate the adhesion and internalization of EVs in target cells [52]. Therefore, both EVs and viruses can be considered as bioactive structures able to influence the cellular behavior.
The presence of multiple similarities between viruses (in particular retroviruses) and EVs, immediately triggered conjecture on the real relationship between vesicles and viruses. For this reason, two alternative theories have been proposed. The first one, called the “Trojan exosome hypothesis”, states that retroviruses are vesicles evolved following a mutation of the gag gene, which was originally encoded by an integrated retro-transposon that directed its expression product towards the route of vesicle generation. In this perspective, the typical characteristics of retroviruses would have been acquired by evolutionary divergence; the pre-existing biogenesis mechanism of vesicle production would have been used to form viral particles [53]. The second theory does not associate viruses to modified exosomes. It justifies the similarities, giving more importance to the phenomenon of convergent evolution, which would lead to the sharing of the same biogenesis pathways for vesicles and viruses [54]. Both theories provide a plausible justification for the affinities observed between viruses and EVs. However, regardless of their possible origin, these affinities certainly have a negative impact on immunological surveillance in the host, since viruses, during infections, can take advantage of these affinities for escaping the immune system by mimicking vesicle composition and behavior [55].
The remarkable resemblance between EVs and viruses has caused quite a few problems in the studies focused on the analysis of EVs released during viral infections. Nowadays, it is an almost impossible mission to separate EVs and viruses by means of canonical vesicle isolation methods, such as differential ultracentrifugation, because they are frequently co-pelleted due to their similar dimension [56,57]. To overcome this problem, different studies have proposed the separation of EVs from virus particles by exploiting their different migration velocity in a density gradient or using the presence of specific markers that distinguish viruses from EVs [56,58,59]. However, to date, a reliable method that can actually guarantee a complete separation does not exist."
Daleč najboljši vir, ki ilustrira celotno virološko farso, je pa tale: https://www.amazon.com/Virus-Mania-COVID-19.../dp/3752629789
Mislim, da bi tole knjigo moral prebrati čisto vsak, tako zelo dobra je.
https://www.amazon.com/Virus-Mania-COVID-19-Hepatitis-Billion-Dollar/dp/3752629789?fbclid=IwAR3YqPcT_z-2E_cT8LjlbHEr_UHhO_zWk-akkZ0zu_6bPvM3JMYin1edQZ0
Pika Dobrovoljec
umiram od smeha. Se ena bedarija. V toksicnem okolju nobene celice ne spravis gor so res cort. Res velikanske budale. Da nebi locil od eksosomov in virusov ha ha ha. Potej teoriji proizvajamo v celicah nove vrste. Uslo mi bo. Ha ha.
Gorazd Janez Pretnar
V znanosti ste že od nekdaj zaprti za nove ideje in tudi danes še vedno vneto verjamete v zastarele dogme, kot je npr. fiksni genom. Jasno je, da se organizmi stalno prilagajamo na okolje in zapis se temu primerno prilagaja. To opisuje tudi zgoraj omenjena knjiga - res zelo priporočam
In ja, pod vplivom stresa (kot je npr. antibiotik, ki ga dodajate celičnim kulturam) se izločajo eksosomi: https://www.nature.com/articles/s41598-017-08392-1
*
Virusi obstajajo: https://animal.mx/2020/05/virus-no-existe-noticia-falsa-stefan-lanka-medicina-germanica/
*
*
https://pubmed.ncbi.nlm.nih.gov/33239378/
Inference of Active Viral Replication in Cases with Sustained Positive Reverse Transcription-PCR Results for SARS-CoV-2
*
Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 Replication and Tropism in the Lungs, Airways, and Vascular Endothelium of Patients With Fatal Coronavirus Disease 2019: An Autopsy Case Series
https://pubmed.ncbi.nlm.nih.gov/33502471/
*
https://pubmed.ncbi.nlm.nih.gov/33330870/
SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome
*
https://science.sciencemag.org/content/sci/370/6513/203.full.pdf
https://science.sciencemag.org/content/370/6513/203.full
In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges
View ORCID ProfileBeata Turoňová1,2,*,
View ORCID ProfileMateusz Sikora3,*,
View ORCID ProfileChristoph Schürmann4,*,
View ORCID ProfileWim J. H. Hagen1,
View ORCID ProfileSonja Welsch5,
View ORCID ProfileFlorian E. C. Blanc3,
View ORCID ProfileSören von Bülow3,
View ORCID ProfileMichael Gecht3,
View ORCID ProfileKatrin Bagola6,
View ORCID ProfileCindy Hörner4,7,
View ORCID ProfileGer van Zandbergen6,8,9,
View ORCID ProfileJonathan Landry10,
View ORCID ProfileNayara Trevisan Doimo de Azevedo10,
View ORCID ProfileShyamal Mosalaganti1,2,
View ORCID ProfileAndre Schwarz1,
View ORCID ProfileRoberto Covino3,11,
View ORCID ProfileMichael D. Mühlebach4,7,
View ORCID ProfileGerhard Hummer3,12,†,
View ORCID ProfileJacomine Krijnse Locker13,†,
View ORCID ProfileMartin Beck1,2,†
See all authors and affiliations
Science 09 Oct 2020:
Vol. 370, Issue 6513, pp. 203-208
DOI: 10.1126/science.abd5223
Flexible spikes
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein enables viral entry into host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor and is a major target for neutralizing antibodies. About 20 to 40 spikes decorate the surface of virions. Turoňová et al. now show that the spike is flexibly connected to the viral surface by three hinges that are well protected by glycosylation sites. The flexibility imparted by these hinges may explain how multiple spikes act in concert to engage onto the flat surface of a host cell.
Science, this issue p. 203
Abstract
The spike protein (S) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is required for cell entry and is the primary focus for vaccine development. In this study, we combined cryo–electron tomography, subtomogram averaging, and molecular dynamics simulations to structurally analyze S in situ. Compared with the recombinant S, the viral S was more heavily glycosylated and occurred mostly in the closed prefusion conformation. We show that the stalk domain of S contains three hinges, giving the head unexpected orientational freedom. We propose that the hinges allow S to scan the host cell surface, shielded from antibodies by an extensive glycan coat. The structure of native S contributes to our understanding of SARS-CoV-2 infection and potentially to the development of safe vaccines.
The spike surface protein (S) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is required to initiate infection (1). It binds to the angiotensin-converting enzyme 2 (ACE2) (2, 3) to mediate viral entry. S also determines tissue and cell tropism. Mutations may alter the host range of the virus and enable the virus to cross species barriers (4, 5). Vaccine efforts focus on neutralizing antibodies that block infection by binding to S.
S is a trimeric class I viral fusion protein (6) with a club-like shape of ~20 nm in length. The ectodomain consists of a head, which has been extensively studied in vitro. It is connected to the membrane by a slender stalk. The three receptor binding domains (RBDs) of the S head are conformationally variable, which may relate to receptor binding. In the closed conformation, the RBDs are shielded by the N-terminal domains (NTDs). In the open conformation, one RBD is exposed upward away from the viral membrane (2, 3). Previous studies resolved roughly two-thirds of the predicted 22 N-linked glycans that are thought to shield S against antibodies (2, 3). It remains unknown whether the distribution of the conformational states and the glycosylation pattern observed with recombinant protein in vitro are representative of the native state generated during viral assembly. Furthermore, little is known about the stalk of S and how its conformational variability within the virion may affect the accessibility of epitopes for neutralizing antibodies and facilitate viral entry.
SARS-CoV-2 virions present prefusion S in an irregular pattern
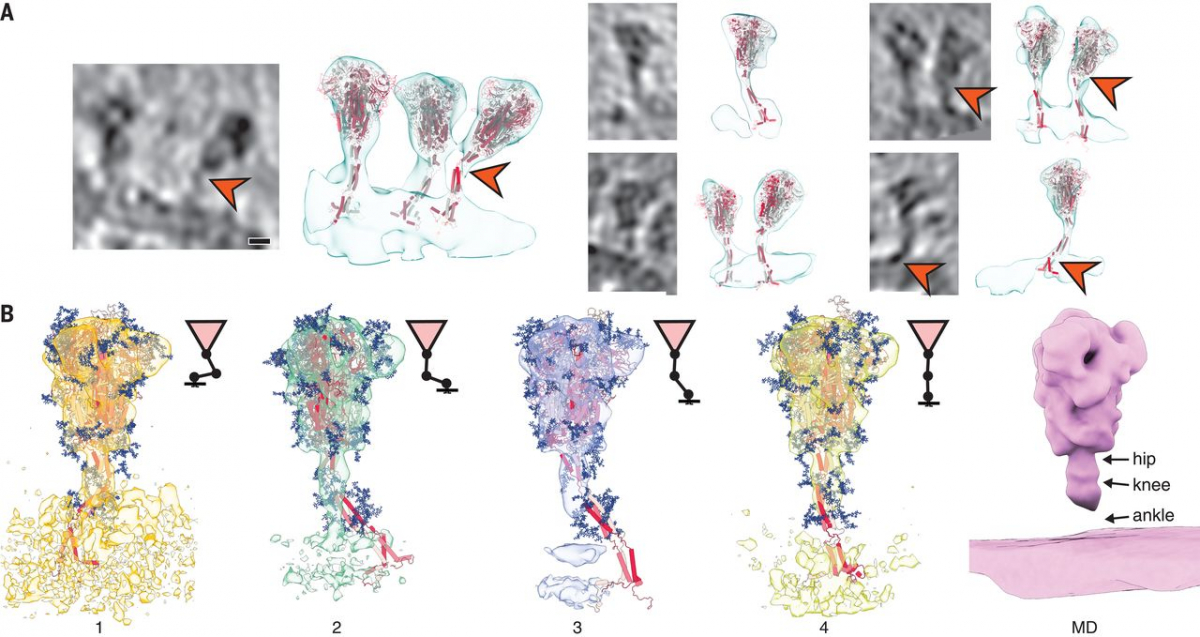
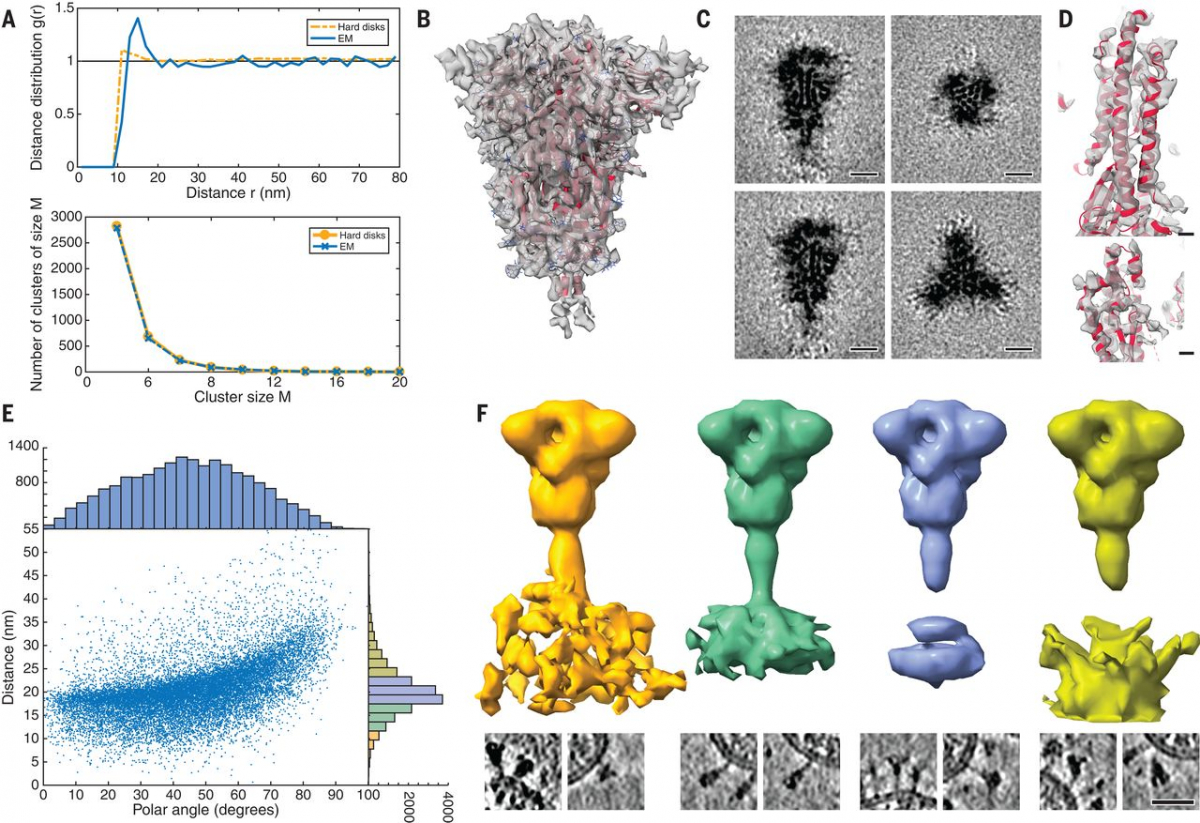
To structurally analyze SARS-CoV-2 S in situ, we passaged the virus through tissue culture cells and used sucrose centrifugation to purify it from the inactivated supernatant (see materials and methods). We acquired a large-scale cryo–electron tomography dataset that consists of 266 tilt series covering >1000 viruses. Visual inspection of the tomographic reconstructions revealed a very-high-quality data set in which individual protein domains were clearly visible (Fig. 1A and movie S1). On average, 40 copies of the S trimer resided on the surface. S proteins appeared to be distributed randomly on the viral surface without any significant tendency to cluster (Fig. 2A).
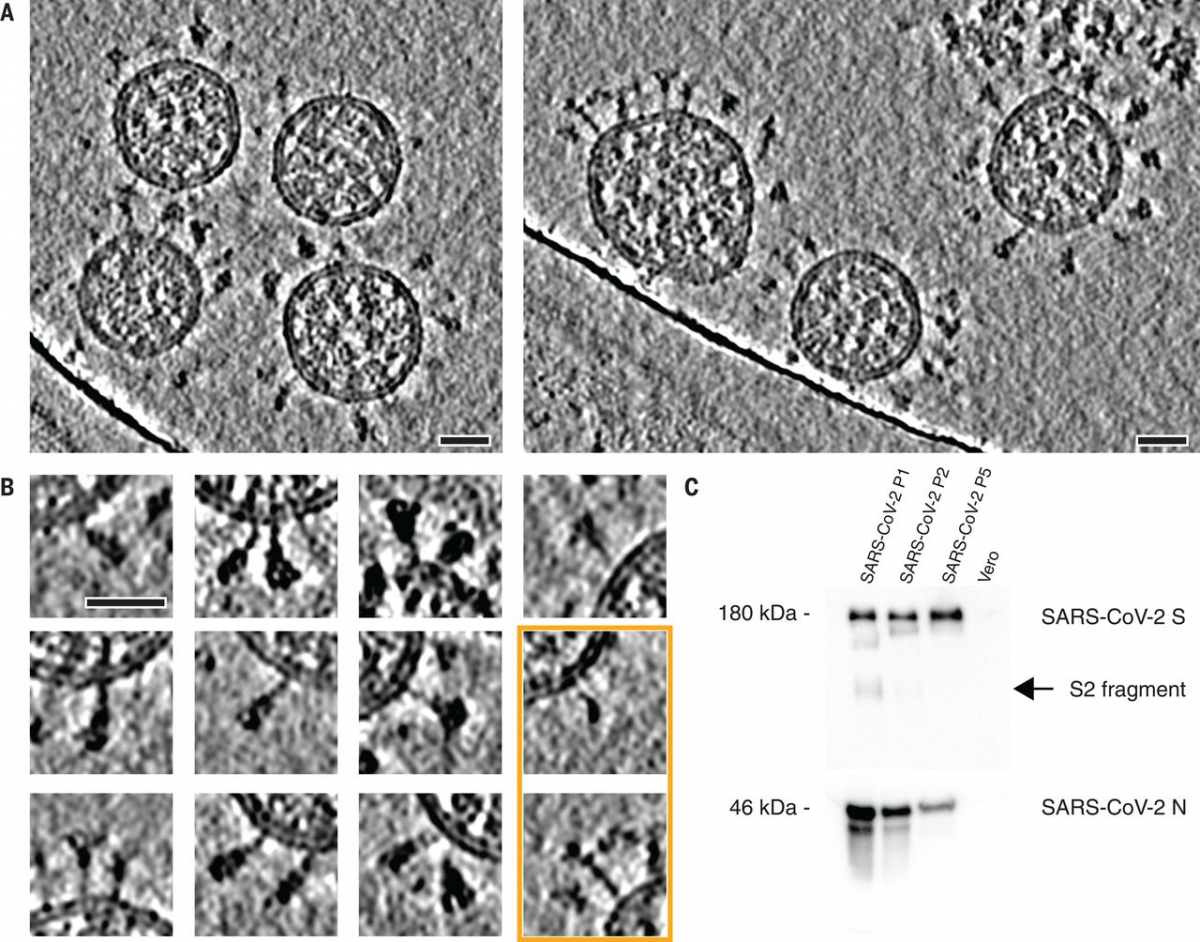
Fig. 1 Cryo–electron tomography of SARS-CoV-2 virions.
(A) Slices through tomographic reconstructions of SARS-CoV-2 virions. Scale bars, 30 nm. (B) Same as (A), but tomograms are arranged as a gallery to highlight specific features of S. All domains, including the transmembrane part, are clearly resolved. Although most of S is reminiscent of the prefusion conformation, the particles framed in orange resemble the postfusion conformation as described by Cai et al. (7). Scale bar, 30 nm. (C) Immunoblot showing the loss of cleavage products of SARS-CoV-2 S with uncleaved S (180 kDa) remaining, within five passages (P1 to P5) through tissue culture (loading control using anti-N antibody). N, nucleocapsid protein.
" data-icon-position="" data-hide-link-title="0" style="box-sizing: inherit; background-color: transparent; color: #37588a; font-weight: bold;">
Fig. 1 Cryo–electron tomography of SARS-CoV-2 virions.
(A) Slices through tomographic reconstructions of SARS-CoV-2 virions. Scale bars, 30 nm. (B) Same as (A), but tomograms are arranged as a gallery to highlight specific features of S. All domains, including the transmembrane part, are clearly resolved. Although most of S is reminiscent of the prefusion conformation, the particles framed in orange resemble the postfusion conformation as described by Cai et al. (7). Scale bar, 30 nm. (C) Immunoblot showing the loss of cleavage products of SARS-CoV-2 S with uncleaved S (180 kDa) remaining, within five passages (P1 to P5) through tissue culture (loading control using anti-N antibody). N, nucleocapsid protein.
Fig. 2 Subtomogram analysis of SARS-CoV-2 S protein.
(A) Distance (top) and cluster-size distributions (bottom) of S on the viral surface, with nonoverlapping hard disks of 10-nm diameter as a reference. (B) Subtomogram average of the ectodomain of S, shown isosurface rendered and fitted with the previously published atomic model as determined by single-particle EM (PDB ID 6VXX). Transparent gray, subtomogram average; red, secondary structure elements; blue, glycosylation sites. (C) Same subtomogram average but shown as slices through the reconstruction. Scale bars, 5 nm. (D) Detail of the average of the symmetric unit of S. Scale bars, 5 Å. (E) Distribution of the angular orientation and distance of the ectodomain with respect to the bilayer. (F) On the basis of the initial subtomogram averaging, positions of the spike head were classified according to their distance from the lipid bilayer (supplementary materials). (Top) Averages of the resulting classes are shown isosurface rendered; distance increases from left to right. (Bottom) Examples of individual particles are shown as slices. At an optimal distance, the stalk domain stretches out and is resolved. Scale bar, 30 nm.
S was mostly present in the prefusion conformation (Fig. 1B). Postfusion conformations (7, 8) were very rare (<0.1%), which appears typical for Vero E6 host cells (9). Sanger sequencing and immunoblot analysis revealed that the furin site for proteolytic cleavage into the S1 and S2 fragments (5, 10) was lost during tissue culture passage (Fig. 1C and fig. S1), consistent with previous studies (11, 12). However, the isolate contained the Asp614→Gly (D614G) allele (13, 14). Large-scale sequencing of RNA isolated from tissue culture supernatant confirmed both findings (supplementary materials).
Subtomogram averaging with NovaSTA (15) and STOPGAP (16) resulted in a cryo–electron microscopy (cryo-EM) map of the S head at 7.9 Å resolution (fig. S2), in which secondary structure elements and individual glycosylation sites were clearly discernible (Fig. 2, B and C). Classification suggested that about half of S was present in the fully closed conformation. A considerable fraction of the remaining subtomograms had one RBD exposed (fig. S3). Structural analysis of the asymmetric unit yielded an average map of the closed conformation at an overall resolution of 4.9 Å. In particular, the cluster of parallel helices in the center of the head was clearly resolved (Fig. 2D and fig. S4).
By contrast, the stalk connecting the S head to the viral membrane appeared to be dynamic. Although the head was fully contained in the tomographic map, only the top of the stalk domain was resolved. Emerging from the neck of the spike head, it contains an 11-residue Leu repeat sequence (L1141, L1145, and L1152) and adopts an unusual right-handed coiled coil, consistent with a recent single-particle structure of the S head (7). We will henceforth refer to this part of the stalk domain as the “upper leg.” Right-handed trimeric coiled coils were long thought to be absent from the structural proteome (17) but can be seen in the postfusion structure of S from the related mouse hepatitis virus (18).
A stalk with three flexible hinges connects S to the viral membrane
The tomographic images suggest the presence of flexible hinges in the stalk. Stalks of individual S proteins are clearly visible in the tomograms (Fig. 1B), but, after averaging, their density declined sharply at the end of the trimeric coiled coil that forms the upper leg (Fig. 2B). Moreover, the head exhibited large positional and orientational freedom. It was tilted up to ~90° with respect to the normal at distances of 5 to 35 nm from the membrane (Fig. 2E). We grouped our subtomograms into four classes, according to their distance from the bilayer, and averaged them separately. At an intermediate distance, parts of the stalk and bilayer were resolved, suggesting a more defined conformation (Fig. 2F). We then subselected ~3200 particles in which the head was oriented roughly perpendicular to the membrane. In the resulting average, the stalk domain was resolved (fig. S5A). Visual inspection of the respective subtomograms, in which the stalk domains are clearly observed, further corroborated the idea of a kinked stalk with potentially several hinges (Fig. 2F). Local refinement of the lower part of the stalk (henceforth referred to as the “lower leg”) resulted in a moderately resolved structure that would be consistent with the continuation of the coiled coil below a flexible hinge (henceforth referred to as the “knee”) (fig. S5B).
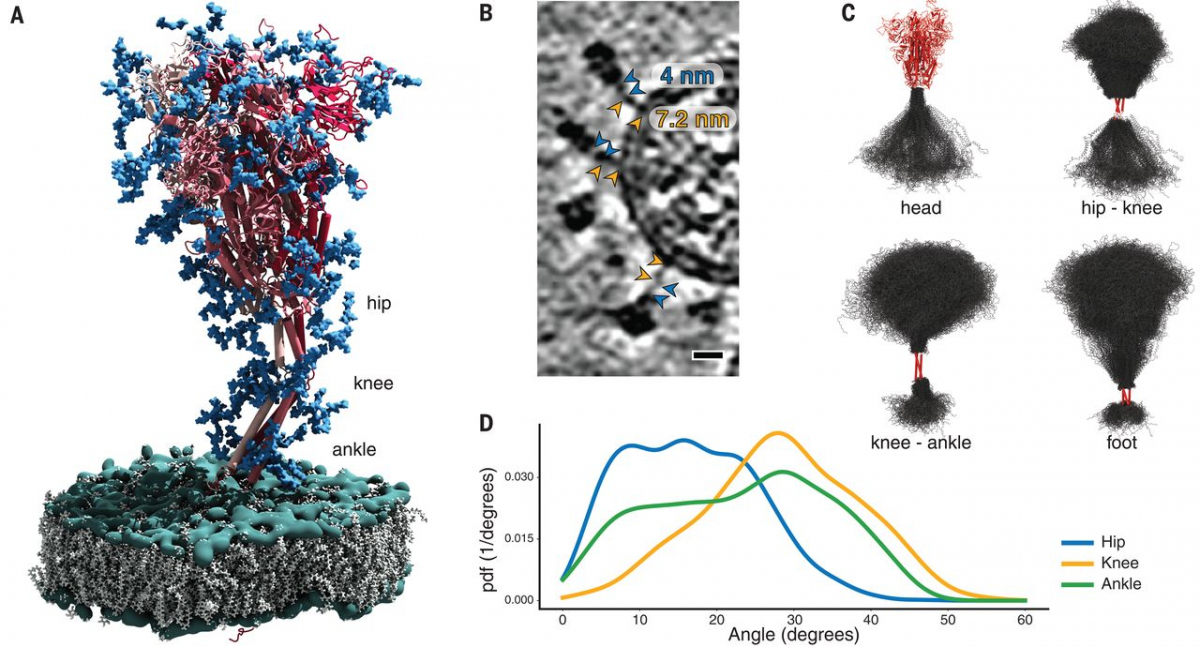
Molecular dynamics (MD) simulations helped us to pinpoint the molecular origins of the flexibility seen in the tomograms. We performed a 2.5-μs-long all-atom MD simulation of a 4.1 million atom system containing four glycosylated S proteins anchored into a patch of viral membrane and embedded in aqueous solvent (Fig. 3A). In the simulations, the S heads remained stable. The stalks, however, exhibited pronounced hinging motions at the junctions between the S head and the upper leg (“hip”), between the upper and lower legs (“knee”), and between the lower leg and the transmembrane domain (“ankle”). This observation was consistent with discrete leg segments seen in the raw tomograms (Fig. 3, B and C). The hip joint flexed the least (16.5° ± 8.8°), followed by the ankle (23.0° ± 11.7°) and the knee (28.4° ± 10.2°) (Fig. 3D and fig. S6). However, the limited sampling in the MD simulation may not have covered the full range of motions (compare Fig. 2E and fig. S6D).
Fig. 3 MD simulations of SARS-CoV-2 S protein.
(A) Model of the S protein. The three individual chains of S are shown in shades of red, N-glycosylation in blue, lipids of the endoplasmic reticulum–like membrane in gray, and phosphates in green. “Hip,” “knee,” and “ankle” mark positions of the three flexible hinges. (B) Examples of the hinges as seen in the deconvoluted tomograms. Blue and orange arrowheads indicate the upper and lower legs, respectively, with their typical lengths indicated. Scale bar, 10 nm. (C) Hinge flexibility in the MD simulation illustrated through backbone traces (gray) at 75-ns intervals with different parts of the S protein fixed (red). (D) Probability density functions (pdf) for hinge bending angles at the hip, knee, and ankle.
Fig. 3 MD simulations of SARS-CoV-2 S protein.
(A) Model of the S protein. The three individual chains of S are shown in shades of red, N-glycosylation in blue, lipids of the endoplasmic reticulum–like membrane in gray, and phosphates in green. “Hip,” “knee,” and “ankle” mark positions of the three flexible hinges. (B) Examples of the hinges as seen in the deconvoluted tomograms. Blue and orange arrowheads indicate the upper and lower legs, respectively, with their typical lengths indicated. Scale bar, 10 nm. (C) Hinge flexibility in the MD simulation illustrated through backbone traces (gray) at 75-ns intervals with different parts of the S protein fixed (red). (D) Probability density functions (pdf) for hinge bending angles at the hip, knee, and ankle.
" data-icon-position="" data-hide-link-title="0" style="box-sizing: inherit; background-color: transparent; color: #37588a; font-weight: bold;">
Structures of S seen along the MD trajectory fit well into the tomographic density of S proteins protruding from the viral surface (Fig. 4A). In particular, the joints of the hip, knee, and ankle of the MD snapshots aligned with kinks in the density visualized by cryo-EM. For a more detailed view, we flexibly fitted suitable snapshots of the MD simulations into the subtomogram averages classified according to the distance of the head from the membrane (compare Fig. 2F to Fig. 4B). Hinge bending gives the S stalk the flexibility required to connect heavily tilted S heads to the viral membrane.
Fig. 4 Fitting of molecular simulations into cryo–electron tomograms.
(A) Slices through tomograms (left) and isosurface-rendered tomograms with snapshots of the respective MD simulations superimposed without flexible fitting (right). The hinges of the stalk domain predicted by structural modeling (orange arrowheads) are consistent with the tomographic data. Scale bar, 5 nm. (B) Fit of snapshots of MD simulations into the classes obtained for different distances of the head from the membrane (1 to 4), as presented in Fig. 2F. Shorter distances are concomitant with a stronger bending of the hinges and a lateral displacement of the stalk. Average MD density filtered to a resolution comparable to the subtomogram averages is shown as an isosurface rendering (right).
Fig. 4 Fitting of molecular simulations into cryo–electron tomograms.
(A) Slices through tomograms (left) and isosurface-rendered tomograms with snapshots of the respective MD simulations superimposed without flexible fitting (right). The hinges of the stalk domain predicted by structural modeling (orange arrowheads) are consistent with the tomographic data. Scale bar, 5 nm. (B) Fit of snapshots of MD simulations into the classes obtained for different distances of the head from the membrane (1 to 4), as presented in Fig. 2F. Shorter distances are concomitant with a stronger bending of the hinges and a lateral displacement of the stalk. Average MD density filtered to a resolution comparable to the subtomogram averages is shown as an isosurface rendering (right).
" data-icon-position="" data-hide-link-title="0" style="box-sizing: inherit; background-color: transparent; color: #37588a; font-weight: bold;">
As a result of hinge bending, the stalk is diluted out in subtomogram averages focused on the head (Figs. 2, B, C, and F, and 4B). Stalks were visible if the heads were aligned with the membrane normal (fig. S5A) or if the stalks themselves were averaged separately (fig. S5B). To test this interpretation, we calculated the electron density averaged over the entire MD trajectory with aligned S heads. Filtered to a comparable resolution, this calculated 3D map was highly similar to the subtomogram averages (Fig. 4B). In rare cases, the coiled coil near the membrane appears to be unfolded in the original tomograms (fig. S5C) and continuous with the disordered loops of the MD model.
Extensive N-glycosylation covers the surface of S
The predicted N-glycosylation sites, many already annotated in single-particle EM maps (2), were generally very pronounced in the subtomogram averages. The electron density of N-glycans averaged over the MD trajectory was highly consistent with the tomographic map (Fig. 5A). Clustered glycosylation sites were visible in the raw density before averaging (e.g., protruding from the lower part of the S head; Fig. 5B). Our analysis of individual sites in subtomogram averages further supports the notion that the spikes were decorated with rather bulky glycan chains (Fig. 5C). Notably, a number of sequons were resolved with more-pronounced branching than previously reported (19). By contrast, the two predicted O-glycosylation sites (20) lacked excess density (fig. S7A). Sequon N17LT, owing to its location on the unstructured N terminus, was not localized in the density (fig. S7B), but elongated features protruding from the tip of the N-terminal domain (fig. S7B) suggested the presence of sequons N74GT and N149KS.
Fig. 5 Analysis of S protein glycosylation sites and epitopes.
(A) N-glycosylation sites are clearly discernible in the subtomogram average of the head. From left to right: Isosurface rendering of subtomogram average with an individual N-glycosylation site indicated (orange arrowhead); superimposed with the MD-calculated density for all annotated N-glycosylation sites; superimposed with previous structural model of the head (PDB ID 6VXX); and superimposed with a snapshot of the MD simulations. N-glycosylation sites are shown in blue. (B) Tomographic slice highlighting an N-glycosylation site (orange arrowheads) in the original data. Scale bar, 5 nm. (C) Highlight of N-glycosylation positions 709 and 1134 of the MD simulations (top) and in a previous structural model (bottom; PDB ID 6VXX, EMDB 21452). The subtomogram average is shown superimposed at different isosurface thresholds (transparent gray). Extensive additional density is visible. (D to F) The stalk domain is heavily glycosylated at the hinges. (D) Example tomographic slices with bulky density at the hinge positions (orange arrowheads). Scale bar, 5 nm. (E) Superposition of the subtomogram averages (transparent gray isosurfaces) of the head (framed red) and the stalk domain (framed green), with a respective snapshot of the MD simulations emphasizing the glycosylation at the hinges. (F) Same as (E) but shown as a maximum intensity projection through the subtomogram averages. Orange arrowheads indicate bulky density at hinges. (G) Fits of snapshots from MD simulations into the surface of a virion. The tomogram is shown isosurface rendered in transparent gray. The position of epitopes for neutralizing antibodies at the RBDs are indicated with blue arrowheads. (H) Cartoon illustrating a hypothetical docking event in which the hinges facilitate the engagement of multiple instances of S with their receptors.
Fig. 5 Analysis of S protein glycosylation sites and epitopes.
(A) N-glycosylation sites are clearly discernible in the subtomogram average of the head. From left to right: Isosurface rendering of subtomogram average with an individual N-glycosylation site indicated (orange arrowhead); superimposed with the MD-calculated density for all annotated N-glycosylation sites; superimposed with previous structural model of the head (PDB ID 6VXX); and superimposed with a snapshot of the MD simulations. N-glycosylation sites are shown in blue. (B) Tomographic slice highlighting an N-glycosylation site (orange arrowheads) in the original data. Scale bar, 5 nm. (C) Highlight of N-glycosylation positions 709 and 1134 of the MD simulations (top) and in a previous structural model (bottom; PDB ID 6VXX, EMDB 21452). The subtomogram average is shown superimposed at different isosurface thresholds (transparent gray). Extensive additional density is visible. (D to F) The stalk domain is heavily glycosylated at the hinges. (D) Example tomographic slices with bulky density at the hinge positions (orange arrowheads). Scale bar, 5 nm. (E) Superposition of the subtomogram averages (transparent gray isosurfaces) of the head (framed red) and the stalk domain (framed green), with a respective snapshot of the MD simulations emphasizing the glycosylation at the hinges. (F) Same as (E) but shown as a maximum intensity projection through the subtomogram averages. Orange arrowheads indicate bulky density at hinges. (G) Fits of snapshots from MD simulations into the surface of a virion. The tomogram is shown isosurface rendered in transparent gray. The position of epitopes for neutralizing antibodies at the RBDs are indicated with blue arrowheads. (H) Cartoon illustrating a hypothetical docking event in which the hinges facilitate the engagement of multiple instances of S with their receptors.
" data-icon-position="" data-hide-link-title="0" style="box-sizing: inherit; background-color: transparent; color: #37588a; font-weight: bold;">
N-glycosylation is also predicted on the knee (N1158HT and N1173AS) and the ankle (N1194ES) in regions not previously resolved by single-particle EM (Fig. 3A). We observed that these positions generally appeared bulkier in tomographic reconstructions than one might expect if they were not glycosylated (Figs. 1B and 5D). Additional density was very clearly observed in subtomogram averages (Fig. 5, E and F, and fig. S5, A and B), and consistent electron density calculated from the MD trajectory aligned on the lower leg (fig. S7C). N-glycosylation in this region of S might protect the functionally important hinges from antibody binding and help to keep them flexible.
Discussion
The two primary structural analysis techniques combined in this study are complementary. Our MD simulations revealed three flexible hinges (hip, knee, and ankle) within the stalk, consistent with the tomographic data. One might speculate that the high degree of conformational freedom of S on the viral surface is important for the mechanical robustness of the virus or may facilitate motions that interfere with antibody access to the stalk. It might also allow S to engage the relatively flat surface of host cells with higher avidity (Fig. 5, G and H). Future tomographic studies of actual infection events might further explore these topics. In contrast to the prefusion conformation of S, the postfusion conformation previously observed in vitro and in situ (7, 9), as well as in this study (Fig. 1B), is apparently inflexible. To the best of our knowledge, extensive flexibility comparable to that of the prefusion S stalk has not been reported for other class I viral fusion proteins, including HIV env, influenza HA, or Ebola GP. However, influenza HA attaches to micelles with a short linker permitting up to 25° bending (21).
A particularly unusual feature masked at the edge of the resolved density of single-particle structures but well resolved in the subtomogram averages is the short right-handed coiled coil at the top of the prefusion stalk. Because this feature is lost in the postfusion structure as resolved for SARS-CoV (8), we speculate that it is only marginally stable, priming the protein for a large structural reorganization in a spring-loaded viral fusion mechanism. Indeed, all three hinges are disassembled in the transition to the postfusion conformation and placed outside the structural core (7, 8).
Overall, the observed distribution of S on the surface of the virion and its conformers is highly consistent with the findings of other studies (9, 22, 23). Host cell-type–dependent differences in the abundance of pre- and postfusion conformation (9, 22) may depend on different levels of ACE2 and the serine protease TMPRSS2 (10). Whether the furin cleavage site plays a role here remains to be addressed. A notable difference is the higher abundance of S on the viral surface observed in this study compared with others (22, 23).
The fully closed prefusion conformation of S was abundant in situ. This finding emphasizes that the highly engineered, recombinant versions of S locked into this conformation (24, 25) may be valuable tools for vaccine development, although there are also differences to the in situ structure. N-glycosylation sites appeared very bulky in the tomographic map compared with previous single-particle analysis, suggesting that decoration with sugars may be more extensive on S assembled in infected cells than on S expressed recombinantly. Our map is suggestive of additional N-glycosylation at the hinges of the stalk domain and possibly on the tips of the S NTDs. The native glycosylation pattern defines the accessibility of epitopes on the crowded viral surface (19), where the NTD and stalk domains appear occluded by neighboring spikes (Fig. 5G). A lack of excess density at the predicted O-glycosylation sites indicates that N-glycosylation dominates.
By using cryo–electron tomography of intact viruses, we were able to resolve functionally important parts of S, including its connection to the viral membrane and its glycan coat, which were masked in studies of recombinant detergent-solubilized protein. Beyond S, our large-scale tomographic dataset contains rich, high-resolution structural information on SARS-CoV-2 particles in their native context. The in situ structures of several key viral components—including the nucleocapsid and the M protein that is highly enriched in the membrane—remain enigmatic. Our data might thus be explored to resolve such features in the future. Furthermore, high-resolution structural models can be fitted directly into the tomographic reconstructions, emphasizing the high quality of the data. This strategy might thus help us to build structural models of entire virions.
Supplementary Materials
science.sciencemag.org/content/370/6513/203/suppl/DC1
*
Virusi NISO eksosomi:
V človeški krvi se resda lahko pojavijo bakterijam podobne celice, na primer zaradi motenj v pH, a to niso virusi ali bakterije ampak skupki beljakovin. Več o tem je v študiji:
https://www.nature.com/articles/s41598-017-10479-8.
Človeška DNK ne vsebuje sekvenc, ki so v genetiki korona retrovirusa SARS CoV-2 in ta ni izloček človeških celic zaradi zastrupitve in NI eksosom. To je povedal velik strokovnjak, ki je tudi s sodelavci napisal študijo o eksosomih:
https://pubmed.ncbi.nlm.nih.gov/22031862/
*
https://link.springer.com/article/10.1007/s12031-020-01767-6 Patogeneza SARS CoV-2
*
SARS-CoV-2 strain Slovenia/SI-4265/20, D614G
Product Description 1ml= 2000 USD
Ref-SKU:
005V-03961
Infectious cell culture supernatant containing SARS-Cov-2 (strain Slovenia/SI-4265/20, D614G). This strain is preserved under Viral Storage Medium -80C. To confirm its identity the virus has been completely sequenced.
Product Risk Group:
ICTV Taxonomy:
Riboviria / Orthornavirae / Pisuviricota / Pisoniviricetes / Nidovirales / Cornidovirineae / Coronaviridae / Orthocoronavirinae / Betacoronavirus / Sarbecovirus / Severe acute respiratory syndrome-related coronavirus
Virus name:
Strain:
strain Slovenia (SI-4265/20), D614G
Storage conditions:
Viral Storage Medium -80C
Sequencing:
Complete genome
Infectivity:
Infectivity tested and quantified
Mycoplasmic content:
Mycoplasma free
GMO:
No
Virus host type:
Virus Cultivability:
Infectivity Test:
TCID50
Viral titer:
10E6.62 TCID50/mL
Production cell line:
Genbank reference:
Passage:
4th passage
Identification technique:
PCR, sequencing,
Shipping conditions:
IATA Classification:
Information about the collection of the virus
Biological material origin:
Natural origin
Collection date: BEFORE
Monday, 27 April, 2020
Country of collection:
Suspected epidemiological origin :
Isolation host:
Human patient; nasopharyngeal swab
*
V Sloveniji glede na podatke portala GISAID, ki ponuja odprt dostop do genomskih podatkov novega koronavirusa, je potrjen primer indijske različice virusa sars-cov-2. Potrdili so jo v Inštitutu za mikrobiologijo in imunologijo medicinske fakultete Univerze v Ljubljani v vzorcu, ki je bil odvzet 20. aprila, je razvidno s portala.
Laboratoriji, ki opravljajo sekvenciranje genomov novega koronavirusa, te podatke vnesejo na portal GISAID. Gre za svetovno znanstveno pobudo in primarni vir, ki ponuja odprt dostop do genomskih podatkov novega koronavirusa.
Predstojnik inštituta Miroslav Petrovec za STA še ni potrdil te informacije. Prav tako z njo niso seznanjeni v Nacionalnem laboratoriju za zdravje, okolje in hrano. V četrtek bodo sicer predvidoma na novinarski konferenci o covidu 19 predstavili stanje glede različic v Sloveniji.
Indijska različica virusa velja za bolj nalezljivo in nevarno in naj bi bila odgovorna za velik porast okužb in smrti zaradi covida 19 v Indiji, kjer iz dneva v dan podirajo črne rekorde.
*
*
SARS‐CoV‐2 AND EXOSOMES
Exosomes are small endosomal‐derived microvesicles secreted by cells to transport biomolecules such as proteins, mRNA, microRNAs, and lipids to the recipient (target) cells. Exosomes are involved in intercellular communications between cells by altering the recipient cell's gene expression and overall function. 125 Exosomes are released both during normal physiological conditions and during pathologies, including viral infections and malignant transformations. 126 , 127 Thus, exosomes have potential to be employed as diagnostic and prognostic molecular biomarkers as well as novel therapeutic modalities. 125
Exosomes have gained popularity during the Covid‐19 pandemic since they are mentioned as one argument by Andrew Kaufman that SARS‐CoV‐2 does not exist. To support his argument, Andrew Kaufman stated that what was detected by PCR is actually not a specific virus, but exosomes. 128 It is clearly seen that this claim is extremely illogical and against common sense since SARS‐CoV‐2 was not solely detected and characterised by PCR but also other modalities, including viral cell culture, whole‐genome sequencing, and electron microscopy technologies.
Indeed, the virus (initially named as 2019‐nCoV) was first isolated from bronchoalveolar‐lavage samples collected on 30 December 2019 by passaging in human airway epithelial cells, Vero E6, and Huh‐7 cell lines. 1 The viral structure of SARS‐CoV‐2 and the cytopathic effect of the infected cells can be clearly visualised by a transmission electron microscopy. Importantly, two nearly full‐length and one full‐length sequences were then submitted and published in GISAID. 1 Subsequently, the genetic sequences of thousands of SARS‐CoV‐2 strains isolated from all over the world have also been published in GISAID, which have an approximate length of 30,000 bases. 129
Interestingly, in his video, Kaufman twisted Dr. James E. K. Hildreth's statement who spoke about HIV in his article, which he quoted as ‘ … the virus is fully an exosome in every sense of the word’ to support his claim that a contagious infectious virus does not exist at all. 128 However, what Dr. Hildreth meant in his paper was that HIV is a virus that hijacks the exosomes in the host cells for both biogenesis of viral particles and viral spread. 130 It is one common mechanism of immune evasion by pathogenic viruses, including hepatitis A virus (HAV) and HCV, since it may facilitate escape from neutralising virus‐specific antibodies. 126 , 127 Clearly, Kaufman intentionally skewed Dr. Hildreth's statement to support his claim that SARS‐CoV‐2 is not a virus, but it is an exosome. 128
Moreover, since SARS‐CoV‐2 is an RNA virus, while exosome can also contain RNA, it is possible to be mistakenly detected, according to Kaufman's statement. 128 Obviously, this is Kaufman's misunderstanding because even though both of them are indeed RNA, the human exosome merely contains human‐derived small RNA (mRNA and microRNA) and cannot release another RNA's species (e.g., virus‐derived small RNA). The human exosome is also unable to release virus‐derived small RNA if the virus itself does not exist in human cells. Exosomes can only release virus‐derived small RNA if the cell has been infected with a virus. 131 Exosomes can also transport fully infectious viral particles, including their genetic material, as has been shown in HCV. 126 Today's technology has been sophisticatedly developed since deep sequencing technology can distinguish between the chain of virus‐ and human‐derived RNA, so it is impossible to mistakenly confuse these clearly different RNA sequences.
Department of Microbiology, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta Indonesia,
2 Center for Child Health—PRO, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta Indonesia,
Mohamad S. Hakim, Email: di.ca.mgu@mikah.s.m.
*
https://www.mcgill.ca/oss/article/covid-19-pseudoscience/psychiatrist-who-calmly-denies-reality
Sounding composed and knowledgeable, Kaufman repeatedly tells his viewers that viruses are not a cause of human diseases. Through watching hours and hours of video, I have seen him deny the existence of the viruses behind the common cold, polio, HIV-AIDS, viral hepatitis, chickenpox, COVID-19, and measles. One of his favourite examples for why his war against germ theory is justified is the case of Stefan Lanka, which he sells to his audience as “the Supreme Court of Germany actually ruled that there is no measles virus that’s been proved to exist” (from his interview with London Real, time code 1:04:00). The truth is that Lanka issued a challenge: he wanted a single scientific paper that, on its own, proved beyond the shadow of a doubt that the virus existed. When a doctor named David Bardens produced six papers that together met the burden of proof, Lanka refused to pay and the Court recognized that Lanka was free to set the rules as he saw fit because this was an award and he could give it to whomever. The measles virus is very real: Lanka’s public challenge was, in my opinion, a no-win scenario to give credence to his virus denialism.
These exosomes can carry payloads, like genetic material, and act as transporters inside our body, and they do look an awful lot like many viruses. In fact, sometimes a virus will infect a cell and an exosome containing the virus’ genetic material will bud off and go on to infect another cell, just like a viral particle would! But here we reach the end of our bridge. Two scientific experts discussed this issue in a YouTube video and concluded that “clearly, there are similarities between exosomes and the coronavirus but they are absolutely different in many aspects.” Kaufman takes a leap and claims the virus does not exist. It’s all exosomes.
In fact, Kaufman loves to mention that doctors who claim to have found an infectious virus have never been able to fulfill Koch’s hallowed postulates. A brief history lesson is warranted. Microbiologist Robert Koch stated during the Victorian era (just before we even really knew what DNA and viruses were) that to prove that a microbe caused disease, you needed to isolate it from living things with the disease and not find it in living things without the disease. And if you took it from a living thing that had it and gave it to a living thing that did not, it should produce disease and you should be able to then isolate this microbe within it. So if scientists have not done this with a particular virus, it gives license to people like Kaufman to claim that we just don’t know.
The problem is that Koch himself realized that requiring his postulates to be fulfilled each and every time was mistaken. He noticed people who were carriers of typhoid fever and of cholera who did not have symptoms. They had the infectious agent but not the disease. Was it proof these microbes did not after all cause the disease? No. Koch’s postulates are historically interesting, but they have essentially been supplanted by guidelines based on the detection of DNA or RNA from the microbe itself.
*
Študija: Exosomi lahko prenašajo mRNA: https://capricor.com/wp-content/uploads/2014/04/2020.11.06.371419v2.full-CAPR.pdf
*
To je za tiste, ki jih zanima, kako so "izolirali" morilski virus. 16. julij 2021
Jerneja, znanstvenica, ki dela v ZDA tudi s PCR testi, piše:
Nekateri se vedno vztrajajo, da so virus izolirali in purificirali (pa ceprav imamo dokument od same UNI LJ, da temu ni tako). Ampak zgleda, kot da ti, ki se to govorijo ne znajo brat
Ko so sekvencirali prvi genom virusa iz pacienta v Wuhanu, zakaj so morali med procesom filtrirat ven sekvence, ki so spadale v cloveski genom? Polovica sestavljenih sekvenc je sla ven in to lepo pise v metodologiji tega clanka https://www.nature.com/articles/s41586-020-2008-3
Naredila sem povzetek tega sekvenciranja tukaj in upam, da bo laikom bolj jasno, zakaj jaz in se veliko nas neidoktriniranih vztrajamo pri tem, da virus ni bil nikoli izoliran in purificiran:
"Uradno so prvič sekvencirali genom novega koronavirusa v laboratoriju profesorja Yong-Zhen Zhang-a in ga objavili 7. januarja 2020 v genski banki (GenBank) pod številko MN908947 [15]. Za sekvenciranje so uporabili RNK material, ki so ga našli v bronhoalveolarnem izpirku 41-letnega bolnika. Bolnik je prišel sedmi dan po začetku simptomov v centralno bolnišnico v Wuhanu na Kitajskem z vročino, oteženim dihanjem in suhim kašljem. Z začetnimi etiološkimi raziskavami so izključili prisotnost virusa gripe in adenovirusov, klamidije (C. pneumoniae) in mikoplazme (M. pneumoniae), ki so le štirje od znanih mikroorganizmov, ki lahko napadajo dihalne organe [16].
Moti nas, da so za izdelavo prve knjižnice, s katero se RNK mikrobov in človeškega RNK prepiše nazaj v stabilnejši DNK in ga namnoži za shranjevanje in vso nadaljno obdelavo v laboratoriju v Shanghaju uporabili orodje, ki je namenjeno za delo s sesalci in ne z virusi (SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian, Takara, Japonska). 150 nukleotidov vsake molekule DNK v knjižnici je bilo sekvenciranih. Dobili so kar 56,5 milijonov odčitkov (reads). Pri sestavljanju na podlagi prekrivanja zaporedij v daljša, ki jim pravimo sestavljena zaporedja (assembled contigs), so uporabili dva različna računalniška programa (Megahit in Trinity). Zaporedja, ki sta jih sestavila programa, med sabo niso bila ekvivalentna, saj je Megahit sestavil nekaj čez 348.000 zaporedij z dolžino od 200 do 30.474 nukleotidov, Trinity pa nekaj čez milijon takih zaporedij z dolžino od 201 do 11.760 nukleotidov. Vsem tem dobljenim zaporedjem so nato odstranil domnevna človeška zaporedja tako, da so jih primerjali z znanimi bazami podatkov za človeški genom. Za veliko sekvenc, ki so v bazi podatkov človeskega genoma, ni znanstveno dokazano, da so prepisane v RNK. Na pomoč nam spet pristopijo računalniški modeli, ki izračunajo, katera sekvenca je lahko prepisana v RNK. Tako so izločili približno polovico neuporabnih človeških zaporedij in določili, da vsa ostala zaporedja pripadajo virusom ali baktarijam. Med preostalimi sta bili tudi najdaljši sekvenci, ki sta ju sestavila Megahit (30.474 nukleotidov) in Trinity (11.760 nukleotidov). Ker sta obe sekvenci zelo podobni celotni sekvenci, ki so jo leta 2018 določili v netopirju (bat SL-CoVZC45; GenBank MG772933), so sklepali, da gre za soroden virus. Za daljšo sekvenco (30.474 nukleotidov) trdijo, da je pokrivala skoraj celoten ‘viralni genom’, ampak končna sekvenca, ki so jo objavili 7. januarja vsebuje le 29.903 nukleotidov. Sprašujemo se, kam je izginil del 571 nukleotidov, ki so razlika med sestavljeno sekvenco v Megahitu in objavljeno sekvenco?
V drugem laboratoriju so sekvencionirali vzorce treh bolnikov, prav tako iz Wuhana. Rezultati so bili konec januarja 2020 objavljeni v članku New England Journal of Medicine (NEJM) [17]. Potek sekvenciranja v tem članku ni podrobno opisan, opisane in objavljene pa so bile tri različne sekvence, za vsakega bolnika posebej. Vse tri sekvence so bile podobne sekvencam MERS-CoV in SARS-CoV. Tako kot pri prvem pacientu se tudi ta študija ni nadaljevala po Kochovih postulatih, s katerimi bi te viruse dejansko dokazali kot povzročitelje pljučnic, in to avtorji jasno napišejo v svojem članku.
Potrebno se je zavedati, da vsa določanja zaporedja nukleotidov, s katerimi naj bi se točno določana vrsta virusa identificirala, slonijo na računalniških modelih. Nikoli ne dobimo celotnega zaporedja virusnega genoma, ampak vedno moramo celotno sliko sestaviti, pri tem pa se poslužujemo verjetnostnega računa. Vse molekule RNK, ki so v vzorcu, se namnožijo v zelo kratke delčke, ki so sekvencirani (150 nukleotidov) in se potem le-te sestavljajo nazaj v prvotne molekule skozi računalniški model. Proces je podoben igranju z Lego kockami. Če hočemo iz Lego kock sestaviti raco, jo bomo sestavili, in to na veliko možnih načinov, samo kocke je potrebno poiskati. Prav tako pa bomo lahko iz njih sestavili ribo."
*
Severe acute respiratory syndrome coronavirus 2 Genome sequencing
*
Demant trditev dr. Kaufmanna, da SARS CoV-2 ne obstaja: https://www.youtube.com/watch?v=MhLGZ3FhLGk
*
Posvet Zdravniške zbornice 29.7. 2021: slika koronavirusa Medicinska fakulteta Ljubljana
*
NIJZ daje povezavo na študije o izlolaciji SARS CoV-2:
*
*
Dr. Gorazd Pretnar: SARS CoV-2 je izoliran: (min 12.00): https://www.facebook.com/stoplaznivimmedijem
*
*
*
»Dokument ni nikakršen dokaz, da virus ne obstaja«
Medicinska fakulteta opozarja na zlorabo njihovega imena ob zatrjevanju neobstoja covida-19. Dokument so zlorabili tudi proticepilski protestniki.
Na medicinski fakulteti opozarjajo, da je dokument o domnevnem neobstoju covida-19, na katerega se sklicujejo tudi proticepilci, ki ne želijo upoštevati pogoja PCT, zloraba. Na to dogajanje je opozoril organizator dogodkov Anton Lamovšek, ki je v današnjem sporočilu STA opozoril, da na prireditvah zaznavajo vse več takšnih obiskovalcev.
Kot je v sporočilu za javnost zapisal direktor King Kong Teatra Anton Lamovšek, so na dogodkih, ki jih organizira, zaznali "več obiskovalcev, ki pogoja PCT ne izpolnjujejo in pri tem mahajo oziroma se sklicujejo na dokument, pod katerim je podpisan dekan medicinske fakultete Igor Švab". Kot piše Lamovšek, imajo s takimi obiskovalci precej težav, saj da so večinoma verbalno agresivni in moteči za samo prireditev.
Na Medicinski fakulteti Univerze v Ljubljani so sicer v objavi na svojih spletnih straneh zagotovili, da njihov dekan Švab ni avtor nobenega izmed dokumentov, ki zanikajo obstoj virusa covid-19.
Kot so pojasnili v objavi na svojih spletnih straneh, se je v preteklem tednu na spletu pojavil posnetek, "v katerem je gibanje Osveščeni prebivalci Slovenije za predstavitev svojih idej zlorabilo ime dekana Medicinske fakultete Igorja Švaba, sklicujoč se na odločbo fakultete s 30. novembra lani". Kot pojasnjujejo na fakulteti, dokument, ki se pojavlja na posnetku, "ni nikakršen dokaz, da virus ne obstaja", temveč v njem trdijo, da niso izvajali znanstvene študije o izolaciji virusa Sars-Cov-2 v skladu s Kochovimi postulati, s čemer so odgovarjali na konkretno vprašanje.
"Ker vprašanje, naslovljeno na fakulteto, sprašuje po strokovno nesmiselnih dokazih, smo odgovorili, da s takimi dokazi ne razpolagamo," so pojasnili.
Po oceni ljubljanske medicinske fakultete gre pri objavi gibanja Osveščeni prebivalci Slovenije za nedopustno zlorabo imena in položaja medicinskega strokovnjaka. "Medicinska fakulteta sprejema in resno jemlje svoje poslanstvo ter sodeluje pri znanstveno podkrepljenem obveščanju laične in strokovne javnosti o vseh bolezenskih stanjih, ne samo covid-19," dodajajo.
In zakaj nimajo dokazov?
Odziv dekana UL Medicinske fakultete, prof. dr. Igorja Švaba dr. med., na objavo v medijih
V preteklem tednu se je na spletu pojavil posnetek, v katerem je gibanje Osveščeni prebivalci Slovenije za predstavitev svojih idej zlorabilo ime dekana UL Medicinske fakultete (UL MF), prof. dr. Igorja Švaba, sklicujoč se na odločbo UL MF dne 30. novembra 2020:Ta dokument, ki se pojavlja na posnetku, ni nikakršen dokaz, da virus ne obstaja.
Gre za dokument, v katerem UL MF trdi, da UL MF ni izvajala znanstvene študije "o izolaciji virusa Sars-Cov-2 v skladu s Kochovimi postulati", ki jo je specificiralo vprašanje. Kochovi postulati, objavljeni leta 1890, so bili narejeni za dokaz bakterijskih okužb, viruse so odkrili kasneje in tudi metode njihovega dokazovanja so povsem drugačne. Kochovi postulati na primer zahtevajo gojenje povzročitelja na sterilnem gojišču, viruse pa je mogoče gojiti samo na celičnih kulturah. Ker torej vprašanje, naslovljeno na UL MF, sprašuje po strokovno nesmiselnih dokazih, je UL MF odgovorila, da s takimi dokazi ne razpolaga.Stališče UL MF je, da je obstoj virusa Sars-Cov-2 nedvomno dokazan s sodobnimi metodami, s katerimi se dokazuje obstoj, prenosljivost in patogenost virusov.UL MF dosledno zagovarja znanstveno utemeljena stališča glede covid-19 in da podpira in spoštuje ukrepe za omejevanje te bolezni, vključno s cepljenjem in pogoji PCT.Dekan UL Medicinske fakultete, prof. dr. Igor Švab, ni avtor nobenega izmed dokumentov, ki zanikajo obstoj virusa SARS-Cov-2 in sporoča, da gre po njegovem mnenju pri objavi gibanja Osveščeni prebivalci Slovenije za predstavitev svojih idej za nedopustno zlorabo imena in položaja medicinskega strokovnjaka. UL Medicinska fakulteta sprejema in resno jemlje svoje poslanstvo ter sodeluje pri znanstveno podkrepljenem obveščanju laične in strokovne javnosti o vseh bolezenskih stanjih, ne samo covid-19. V preteklih mesecih smo tako organizirali številne javnosti dostopne seminarje, na katerih so znanstveniki odgovarjali na odprta vprašanja ter pojasnjevali široko področje bolezni covid-19. Vsa gradiva so še vedno dostopna javnosti in priporočamo ogled vseh vsebin: https://lifelong.mf.uni-lj.si/course/index.php?categoryid=29
O izolaciji virusa ...
Since being identified in Wuhan, China in December 2019, the SARS-CoV-2 virus has led to more than 51 million cases of COVID-19 worldwide and over 1.2 million deaths, according to the World Health Organization’s tally from November 12.
Research shows the virus was isolated early in the outbreak and has since been isolated many times in multiple countries.
An article in the Bulletin of the World Health Organization said researchers first isolated the virus in December 2019, and in February scientists analysed the genome from 10,022 samples to understand its variability.
An article published in the journal Nature in February detailed how SARS-CoV-2, a novel coronavirus, was isolated by scientists in Wuhan.
It said full-length, near-identical genome sequences for the virus were obtained from five patients at an early stage of “an epidemic of acute respiratory syndrome” that began in the Chinese city on December 12.
The US Centers for Disease Control and Prevention (CDC) said SARS-CoV-2 had been isolated in its lab and had been available for free to researchers and scientists to study since February.
An article in the CDC journal, Emerging Infectious Diseases, detailed how US scientists isolated SARS-CoV-2 from a patient in January.
Journal articles also detail how the virus was isolated in Korea and Germany.
In Australia, it was isolated from the first person in the country diagnosed with COVID-19, a Wuhan man who was admitted to a Melbourne hospital with fever, cough and laboured breathing.
The University of Melbourne explained in January how scientists in the city had been the first to grow the virus in a lab outside China.
Pathology professor Karen Mossman, part of the team that isolated the virus in Canada, wrote in an article on The Conversation that isolating the virus required collecting specimens from patients and culturing, or growing, it by getting it to infect live mammalian cells in a lab.
A blog post by virologist and adjunct associate professor at the University of Queensland Ian M Mackay also detailed a number of instances where the virus had been isolated around the globe and debunked some of the misconceptions that the virus had not been isolated.
The virus had also been successfully isolated from urine and from ocular secretions.
University of Auckland associate professor and microbiologist Siouxsie Wiles said the theory that SARS-CoV-2 had not been isolated was spreading based on government information requests worded in ways that misunderstood how the isolation process and viruses worked.
In the extended video posted on Instagram, Ms O’Doherty discusses an information request which asked the UK health department for records that showed SARS-CoV-2 had been isolated “from a symptomatic patient of COVID-19 where the sample was not first combined with any other source of genetic material”.
Dr Wiles said that was not possible – describing the request as “a problem of definitions”.
“Viruses are basically inanimate objects which need a culture to activate in. But the way they are phrasing the requests is that the sample must be completely unadulterated and not be grown in any culture – and you can’t do that,” she told in a phone interview.
“You can’t isolate a virus without using a cell culture, so by using their definition it hasn’t been isolated. But it has been isolated and cultivated using a cell culture multiple times all around the world.”
University of Auckland senior virology lecturer John Taylor told that SARS-CoV-2 had been isolated many times using cells to amplify the virus from patient samples and the idea it could represent some kind of contamination originating in the cells was nonsense.
“Viruses are obligate parasites, which means that they only grow within a living host cell. So if you have the caveat that you can’t combine the virus with a host cell then you get the answer you want by ignoring the fundamental biology,” he said.
University of Auckland vaccinologist and associate professor Helen Petousis-Harris said SARS-CoV-2 had been isolated – and it could even be viewed with a powerful microscope.
“We know what it looks like, we’ve got the architecture of it; for example, you can see the spike proteins under a microscope which looks like a halo, or a corona, which is why it’s called a coronavirus.
“You can’t grow a virus without a cell culture, it can’t replicate without the cell machinery, but you can recreate the virus from its genome.”
*
https://www.mdpi.com/2075-4418/11/7/1270/htm
Laboratory Diagnosis of SARS-CoV-2 Pneumonia
*
https://www.technocracy.news/mercola-yes-sars-cov-2-is-real-virus/
*
There are an estimated 1.67 million viruses yet to be identified that currently infect mammals and birds. Of these, it is thought that up to 827,000 have the potential to infect humans.
*
*
Na asteroidu so našli sekvence RNA: https://www.sciencealert.com/scientists-discover-rna-component-buried-in-the-dust-of-an-asteroid?utm_source=ScienceAlert+-+Daily+Email+Updates&utm_campaign=2aeb2878d1-RSS_EMAIL_CAMPAIGN&utm_medium=email&utm_term=0_fe5632fb09-2aeb2878d1-366127361
Nov 23, 2020
